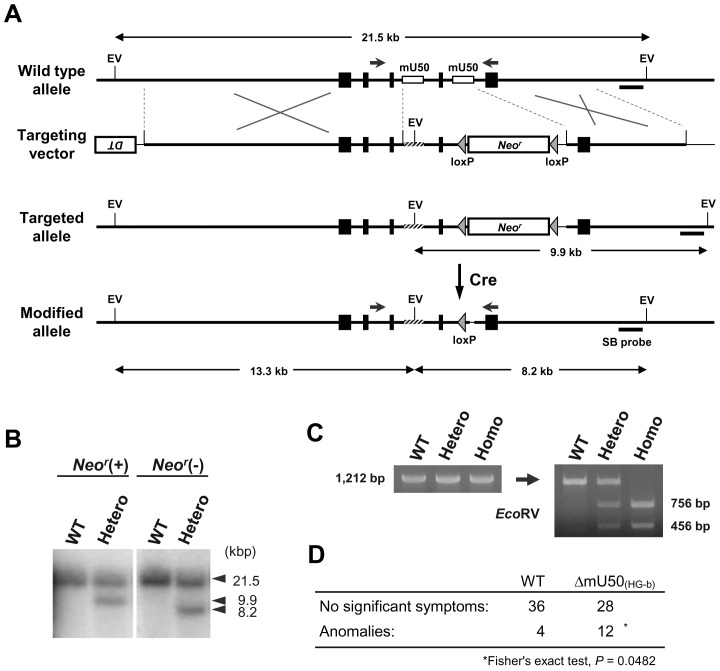
Figure 2. Generation of the mU50-deficient mice.
(A) Schematic representation of the targeting strategy based on the wild-type allele. In the targeting vector, the first mU50 snoRNA sequence (upstream) is substituted by the 68 bp multiple cloning site of a pBluescriptII KSII plasmid containing an EcoRV recognition site (EV). The second mU50 snoRNA sequence (downstream) is replaced with a loxP-Neor-loxP cassette. The pair of facing arrows indicates the set of forward/reverse primers used for genotyping PCR. DT: diphtheria toxin negative selection cassette; Neor: neomycin resistant gene cassette; Cre: treatment with Cre recombinase; SB probe: a probe for Southern blot. (B) Southern blot of genomic DNA from mutant heterozygotes before (left) and after (right) removal of the Neor cassette by Cre recombinase. WT: wild-type; Hetero: mutant heterozygotes. (C) Genotyping PCR for the mU50HG-b gene. Note that all the PCR products had nucleotide lengths that were identical to that of the wild-type. After digestion of the PCR product with EcoRV, fragmentation of the mutant allele-derived amplicon was observed. WT: wild-type; Hetero: mutant heterozygotes; Homo: ΔmU50(HG-b) mutant. (D) Lifelong monitoring of the health condition (n = 40 per genotype) of ΔmU50(HG-b) and wild-type mice. Note the greater number of anomalies in the ΔmU50(HG-b) population. Detailed list is available in Table S1.