Figure 2.
Triggers and Processing Mechanisms of PhasiRNAs
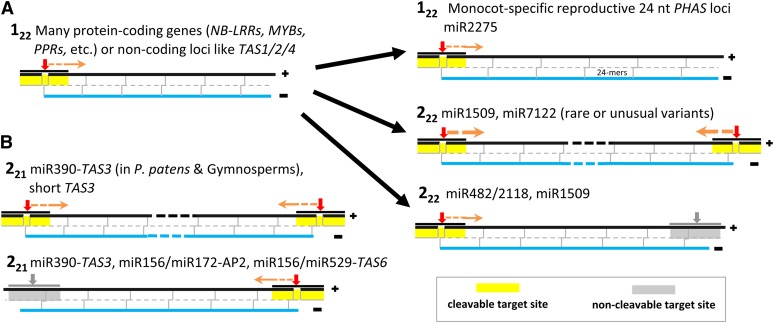
The primary mechanisms of processing for plant phasiRNAs are described along with prototypical loci and the miRNAs that trigger siRNA biogenesis at these loci. Red arrows indicate cleavage sites, and orange arrows indicate the direction of precursor processing into phasiRNAs, which are indicated by gray lines in the double-stranded black/blue precursors.
(A) The one-hit pathway is typified by a single target site for a 22-nucleotide miRNA that results in downstream processing of the target transcript into ∼21-nucleotide phasiRNAs. This is denoted as a 122 locus. There are at least three notable variations on the one-hit model, including (1) the reproductive lncRNAs of monocots that are processed by DCL5 into 24-nucleotide phasiRNAs, triggered by miR2275 and thus also 122 loci, but with different biogenesis components; (2) and (3) are both 222 loci, but the 3′ site can be either cleaved or not cleaved.
(B) The two-hit pathway is typified by two target sites of a 21-nucleotide miRNA that results in processing upstream of the 3′ site. This is denoted as a 221 locus, and the best characterized examples are TAS3 and related loci, although a few other examples have been described. The 5′ site may be cleaved, which may result in processing from both directions, or the 5′ site may be noncleaved, as originally described for the Arabidopsis TAS3 locus.