Figure 4.
miRNAs Target Conserved Sequences in Members of the NB-LRR Gene Families.
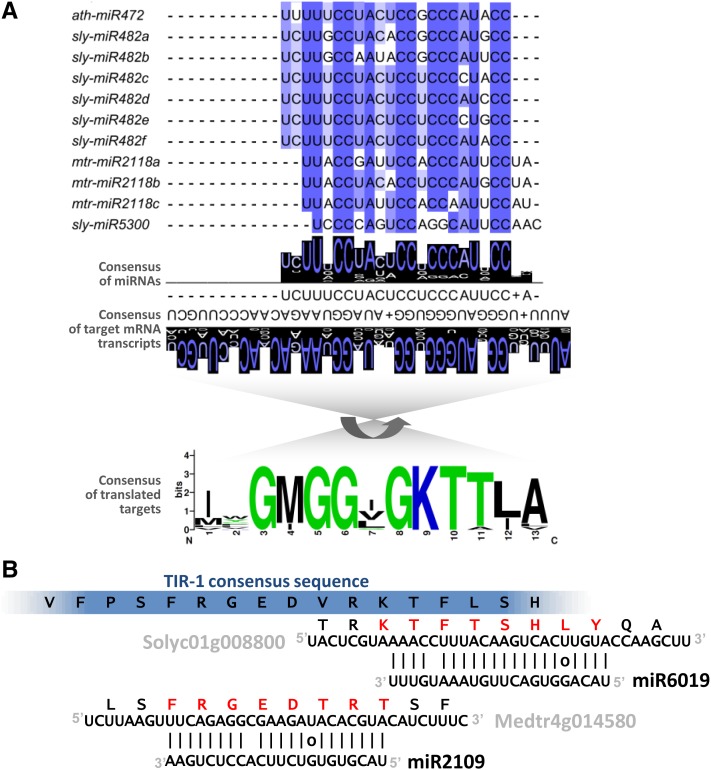
(A) The miR482/miR2118 superfamily of miRNAs is a relatively diverse group (above) that typically targets nucleotides encoding the P-loop motif of NB-LRR proteins (below). Consensus sequences of either the miRNAs or their targets demonstrate a high degree of conservation (illustrated by WebLogo). In this figure, for illustrative purposes, we’ve randomly selected a diverse set of miRNA superfamily members (listed by their names in miRBase) and ∼16 targets from the same source species. In this case, the targets were predominantly CNLs, but miR2118 also targets many TNLs. For ease of alignment, the miRNAs are shown 5′ to 3′ in the consensus, the mRNA targets are shown 3′ to 5′, and the translated protein motif is inverted relative to the target mRNAs (indicated by the curved arrow).
(B) Redundancy in miRNA targeting at the encoded TIR-1 motif of NB-LRRs. miR6019 and miR2109 aligned to their TNL-encoding targets show they target the same encoded motif (blue bar at top), but at adjacent, nonoverlapping sites. The consensus at the top is from Meyers et al. (1999). Red letters indicate the amino acids encoded by the target region. Example targets from tomato (“Soly…”) and Medicago (“Medtr…”) are indicated aligned to the miRNA sequences.