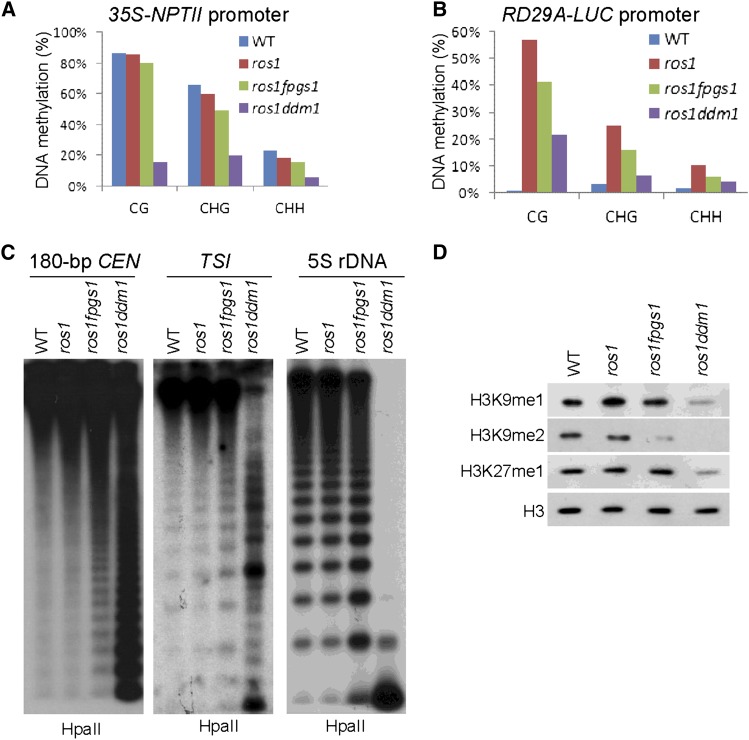
Figure 2.
The Effect of fpgs1 and ddm1 on DNA and Histone Methylation.
(A) and (B) Detection of DNA methylation at the promoters of 35S-NPTII and RD29A-LUC transgenes by bisulfite sequencing. The DNA methylation levels at CG, CHG, and CHH sites are shown. WT, the wild type.
(C) The DNA methylation of 180-bp centromeric DNA, TSI, and 5S rDNA was evaluated by DNA gel blotting. Genomic DNA was cleaved by the DNA methylation-sensitive endonuclease HpaII followed by DNA gel blotting.
(D) The overall levels of H3K9me1, H3K9me2, and H3K27me1 determined by immunoblotting. The protein extracts were isolated from the wild type, ros1, ros1 fpgs1, and ros1 ddm1. The H3 signal was detected as a loading control.