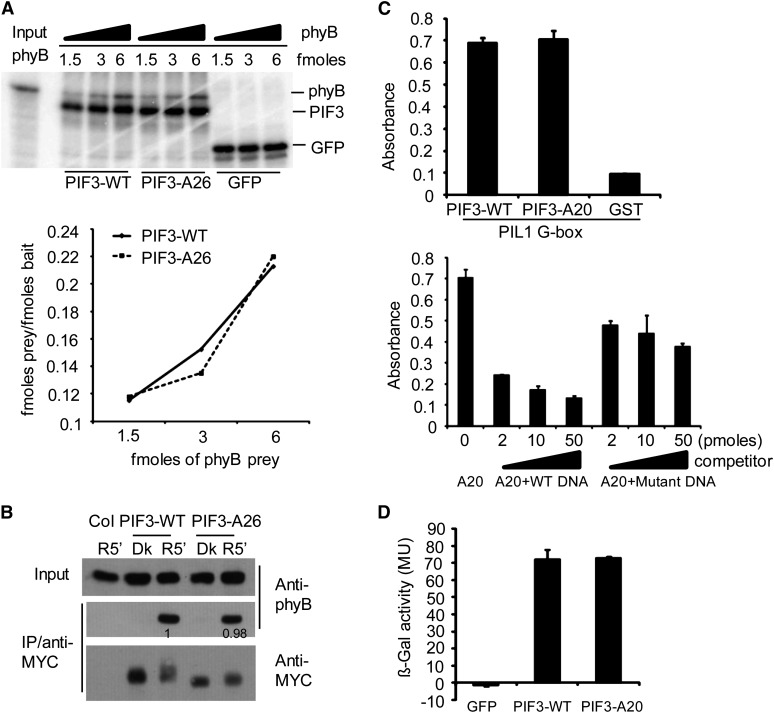
Figure 3.
Multisite Mutations at the Light-Induced PIF3 Phosphorylation Sites Do Not Affect Its Binding Affinity for the phyB Protein, Recognition of a Target Gene Promoter in Vitro, or Transcriptional Activation Activity in Yeast.
(A) PIF3-WT and PIF3-A26 have similar binding affinity for Pfr phyB in vitro. Top panel: Autoradiographs showing interactions of PIF3 with different fold dilutions of phyB prey. All proteins were produced in a TNT in vitro expression system labeled with [35S]Met. PIF3-WT, PIF3-A26, and GFP were first immunoprecipitated with an antibody against the MYC epitope present in each of these constructs and then used as baits to coimmunoprecipitate Pfr phyB. Bottom panel: Quantitative analysis of the data obtained in the top panel. The amount of each bait and prey used was calculated from a standard curve using a known amount of [35S]Met. The femtomoles of prey/femtomoles of bait are plotted against increasing amount of the phyB prey used.
(B) PIF3-WT and PIF3-A26 have similar conformer-specific, apparent binding affinity for Pfr phyB upon coextraction from R light–irradiated seedlings. Proteins were extracted from dark-grown transgenic seedlings that had been irradiated (R5′) or not (Dk) with R light for 5 min. Transgenically expressed MYC epitope–tagged PIF3 was immunoprecipitated (IP) with goat polyclonal MYC antibody and subjected to immunoblot analysis using monoclonal MYC (PIF3 bait) or phyB (prey) antibody. Proteins from nontransgenic Col seedlings were used as the negative control. Numbers directly on the blot image denote the relative phyB protein levels normalized to the PIF3:MYC bait.
(C) PIF3-WT and PIF3-A20 show similar apparent binding affinity for a G-box DNA sequence motif from the PIL1 gene promoter in vitro using a DPI-ELISA assay. Top panel: In vitro binding assay with a PIL1 G-box DNA probe using recombinant GST:PIF3:WT or GST:PIF3:A20 proteins, and GST protein as a negative control. Bottom panel: Competition binding assay of GST:PIF3-A20 with different concentrations of wild-type or mutant G-box probes. Data represent the means of independent duplicates ± se. WT DNA, wild-type competitor probes; mutant DNA, competitor probes mutated in the G-box motif at positions known to eliminate sequence-specific binding by the PIFs.
(D) PIF3-WT and PIF3-A20 have similar transcriptional activation activity in yeast. PIF3-WT and PIF3-A20 were fused to the LexA DNA binding domain and tested for autonomous transcriptional activation activity in yeast using a standard liquid ONPG assay. A GFP fusion protein was used as negative control. MU, miller units. Data represent the means of independent duplicates ± se.