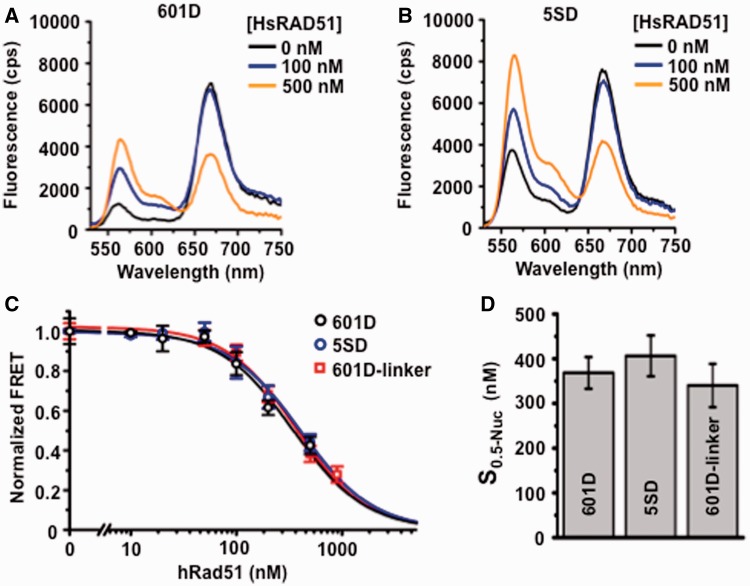
Figure 3.
Nucleosome disassembly is independent of linker DNA and nucleosome positioning stability. (A) Representative fluorescence emission spectra of 601D nucleosomes (12 nM) incubated with HsRAD51 at 0 (black), 100 (blue) and 500 nM (orange) in the presence of 130 mM KCl, ATP (250 µM) and 2 mM Mg2+. (B) Representative fluorescence emission spectra of 5SD-linker nucleosomes (12 nM) incubated with HsRAD51 at 0 (black), 100 (blue), 200 (green) and 500 nM (orange) in the presence of 130 mM KCl and 2 mM Mg2+. (C) Normalized FRET efficiency of 601D (black open circle) and 5SD (black open circle) nucleosomes compared with 601D-linker nucleosomes (black open square) as a function of HsRAD51 concentration (see text). (D) Concentration of HsRAD51 (nM) at which 50% of nucleosomes are disassembled (S0.5). Values represent the average and standard of deviation from at least three independent experiments.