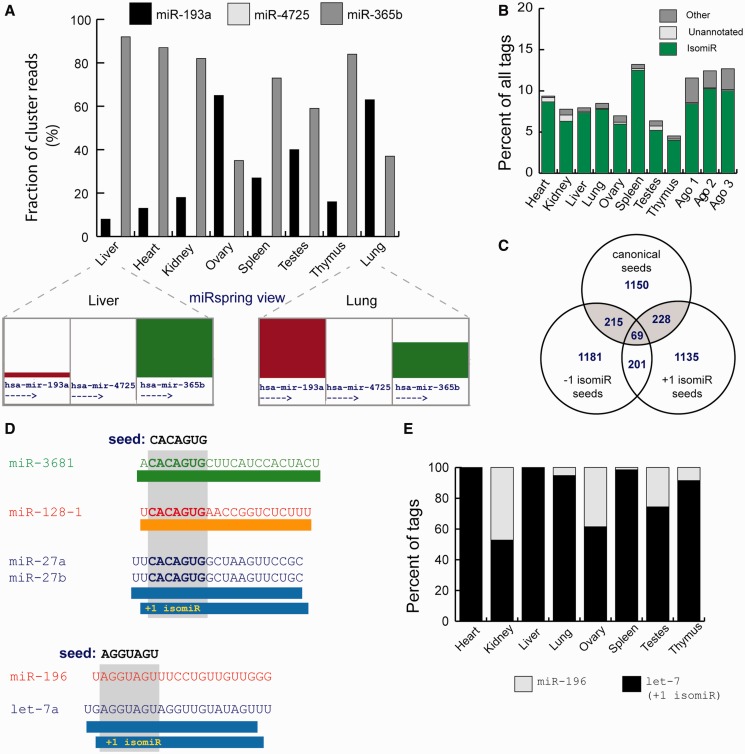
Figure 3.
Analysis of published data sets using the miRspring document. (A) Differential expression or processing of miRNA precursors within a miRNA cluster across different human tissues. (B) Proportion of non-miRBase defined seeds identified in human tissues. Majority are derived from isomiRs of defined-processed miRNAs, and the remainder is derived from miRNAs that are processed from miRBase precursor arms that in miRBase v19 have no defined mature sequence (unannotated) or other processed sequences. (C) Vennn diagram showing the number of miRNAs and their isomiRs that have identical seeds. Seed-isomiRs are highlighted in gray. (D) Examples of interesting seed-isomiRs. The +1 isomiR of miR-27b has the same seed as miR-128 and miR-3681. Similarly, the +1 isomiR derived from the let-7 family has the same seed as miR-196 family. (E) The proportion of miRNAs and their isomiRs that have the canonical miR-196 seed (AGGUAGU) in different human tissues.