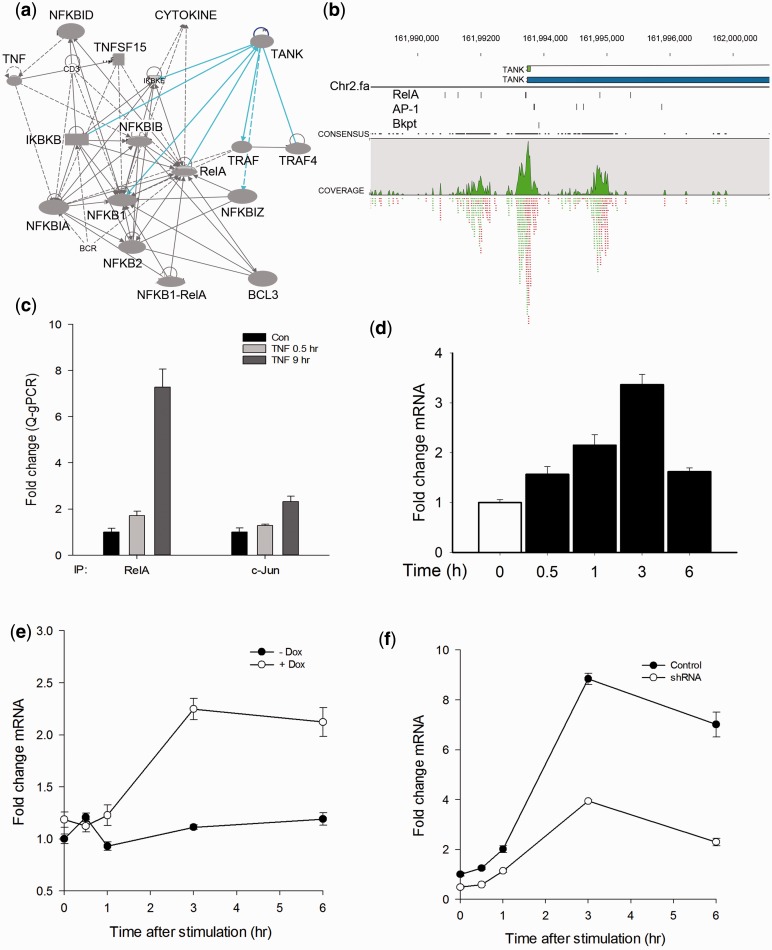
Figure 6.
Validation of TANK as an NF-κB/RelA target. (A) Ingenuity pathway network of NF-κB family members identified in the 20% of significant peaks. Each node is a gene; edges indicate protein–protein interactions (solid lines) and activation (directed arrow). Abbreviations are BCL3, B cell CLL/lymphoma 3; BCR, B-cell receptor; NFKID, IκB-Δ; NFKBIZ, IκB-ζ; TNFSF15, TNF secreted ligand 15 kDa; NFKBIA, IκBα, IKBKB, Inhibitor of IκB kinase β; IKBKE, IκBε; TANK, TRAF family member associated NF-κB activator; TRAF, TNF-associated factor. (B) Top panel, location of NF-κB/RelA peaks on the TANK gene. Shown are TANK gene 5′-UTR, TSS and TANK exons as small green boxes. Bottom panel, NF-κB/RelA peaks (green). (C) Validation of NF-κB/RelA and AP1 binding. XChIP was performed on control or TNF-stimulated A549 cells. Chromatin was immunoprecipitated using RelA or AP1 Abs as indicated. Shown is fold enrichment of TANK promoter sequences by Q-gPCR. (D) Induction of TANK mRNA expression. A549 cells were stimulated with TNF, and TANK mRNA quantified by Q-RT-PCR. Shown is fold change in normalized TANK mRNA as a function of time. (E) Validation of NF-κB dependence on TANK expression. HeLatTA/FLAG-IκBα Mut cells were plated in parallel in the absence (−Dox) or presence (+Dox) of doxycyline (Dox, 2 μg/ml) and stimulated with TNF. TANK mRNA abundance was determined by Q-RT-PCR. (F) AP1 knockdown effect on TANK expression. A549 cells were transfected by lentiviral control shRNA (pLOK.1) or c-Jun shRNA and the pooled puromycin-resistant cells were further treated by TNF and analyzed TANK expression by Q-RT-PCR. Shown is normalized fold change mRNA expression.