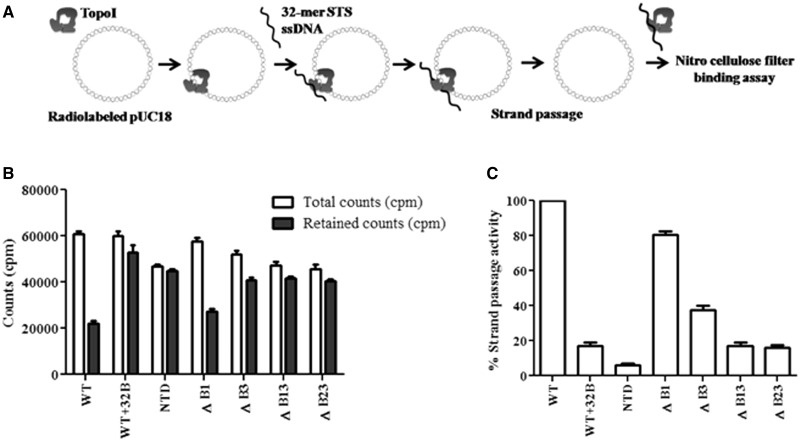
Figure 2.
Assessment of strand passage. (A) Experimental design of filter-binding assay. Nick translated pUC18 DNA was incubated with 500 nM proteins at 37°C. The 32-mer oligonucleotide harboring STS was added to the enzyme–DNA complex to form cleaved DNA gate for the passage of pUC18 DNA. Radioactive counts (counts per minute) associated with each reaction were measured by filter binding assay; (B) Retention of pUC18 DNA. Graph showing the amount of total count and retained count associated with the enzymes. Total counts represent the counts (counts per minute) associated with the enzyme–pUC18 DNA complex. Retained counts (cpm) indicate remaining counts after the completion of strand passage reaction. WT + 32B represents the negative control in which 32B oligonucleotide was annealed with 32 mer and duplex DNA added to enzyme-pUC18 DNA complex. NTD alone used as a control did not show significant difference, suggesting that the decrease in the count is mediated by CTD; (C) Strand passage activity. Graph representing the strand passage activity of the WT and mutant enzymes. Strand passage activity of WT enzyme was normalized to 100%. The error bars represent standard deviation across three measurements. The plasmid DNA molecules, which entered the enzyme cavity, were captured by using a clamp closing anti-topoI monoclonal antibody, and the weakly bound salt sensitive complexes were removed by washing with the high salt buffer. The radioactive counts represent the salt resistant enzyme–DNA hetero-catenanes.