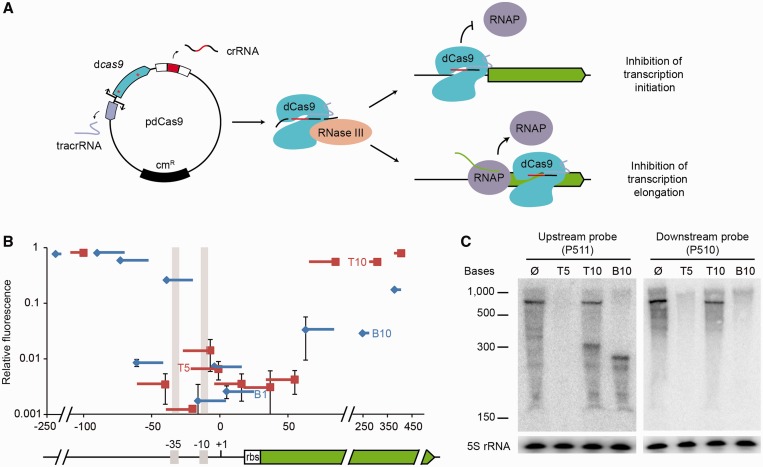
Figure 1.
dCas9-mediated repression in E. coli. (A) Plasmid pdCas9 encodes a cas9 mutant containing D10A and H840A substitutions (red asterisks) that abrogate nuclease activity. dCas9 binds to a tracrRNA:precursor crRNA and recruits RNase III to process the precursor and liberate the crRNA. The crRNA directs binding of dCas9 to promoter or open reading frame regions to prevent RNAP binding or elongation, respectively. (B) GFP fluorescence of cells expressing dCas9 guided to different regions of the gfp-mut2 gene, relative to the fluorescence of cells expressing a non-targeting dCas9, as a function of the position of the target sequence within the gene (+1, transcription start). Squares indicate the PAM position, lines the extension of complementarity between the crRNA guide and the reporter gene. Red and blue lines indicate crRNAs sequences identical to top or bottom DNA strand, respectively. Error bars show one standard deviation from the mean of three relative fluorescence values. The gfp-mut2 gene (green), its promoter, including the −35 and −10 elements (gray shade) and the ribosome binding site (rbs) are shown as reference for the localization of the dCas9 binding sites. (C) Nothern blot with probes annealing either upstream or downstream of the T10 and B10 target sites using RNA extracted from cells expressing T5-, T10-, B10-guided dCas9 or a control strain without a target. Detection of 5S RNA serves as control.