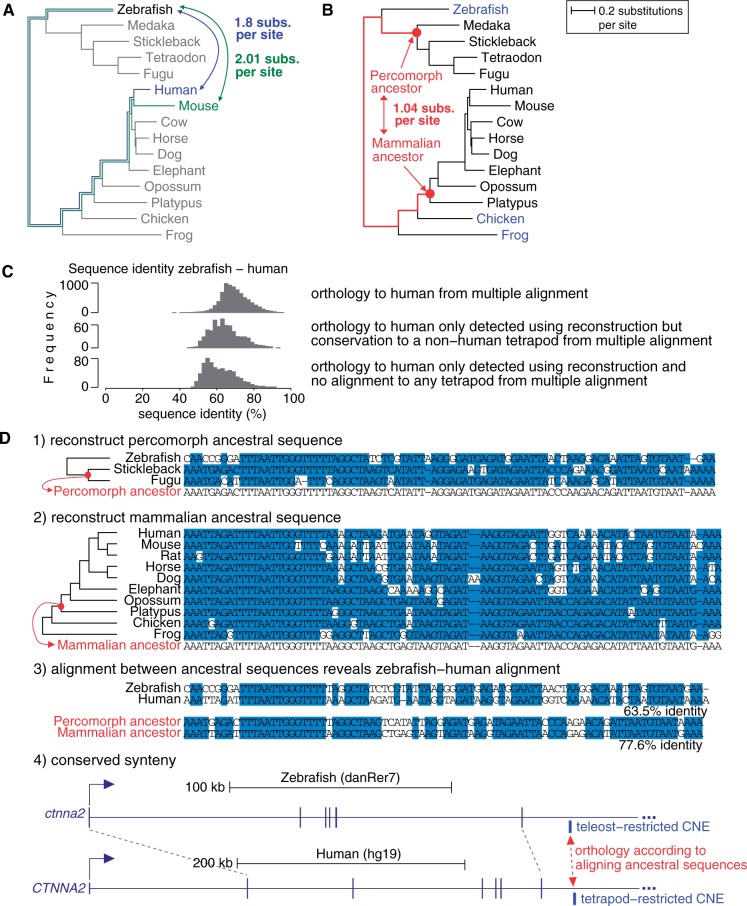
Figure 4.
Ancestral reconstruction reveals additional CNE alignments between distant species. (A) Large evolutionary distances between zebrafish and human/mouse can be substantially reduced if (B) reconstructed ancestral sequences are aligned. The phylogenetic tree contains the species used to reconstruct the percomorph and mammalian ancestor. Species used as outgroups are in blue in (B). (C) Sequence identity of zebrafish–human alignments is shown for CNEs that align to human in our multiple alignment and for 1262 CNEs where ancestral reconstruction but not direct alignment detects conservation to human (630 align to a tetrapod but not human in our multiple alignment; 632 have no alignment to any vertebrate). Although alignments detected only using reconstruction have lower sequence identities, even values ∼50% indicate clear conservation between species separated by ≥1.8 neutral substitutions per site. (D) An example where conservation within teleosts and within tetrapods can be used to reconstruct the percomorph and mammalian ancestor of the CNE (1 and 2). The reconstructed ancestral sequences align with high enough sequence identity to detect orthology and anchor an alignment between the human and zebrafish CNEs not visible otherwise (3). The CNE shares conserved synteny with the same putative target gene (4). Blue background is identity to the ancestor in (1 and 2) and sequence identity in (3).