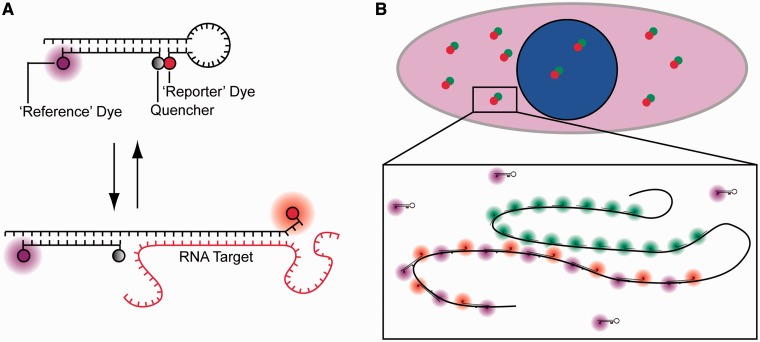
Figure 1.
Schematic of RBMBs and the methodology used to assess RBMB performance in cells. (A) RBMBs are hairpin-forming oligonucleotide probes that are labeled with a reporter dye, quencher and reference dye. The reporter dye is held in close proximity to the reference dye in the absence of target, leading to a low fluorescent state. Hybridization of the loop-domain to complementary RNA causes the fluorescent dye and quencher to separate, resulting in the restoration of fluorescence. The reference dye remains unquenched regardless of the conformation of the RBMB. The double-stranded domain with a 3′-UU overhang drives nuclear export. (B) To evaluate the ability of RBMBs to quantify the number of RNA transcripts in single cells, cells are fixed following the delivery of RBMBs and single-molecule FISH is performed. FISH probes are represented in the schematic as short linear oligonucleotides with a green fluorescent dye, while RBMBs are drawn with a red reporter dye and magenta reference dye. The large number of RBMBs and FISH probes that bind to each RNA transcript results in bright punctate spots within cells, under fluorescence microscopy. Colocalization of RBMB-signal with the smFISH-signal indicates that the RNA transcript was successfully detected by RBMBs.