Fig 3. CAD as a direct substrate of S6K1.
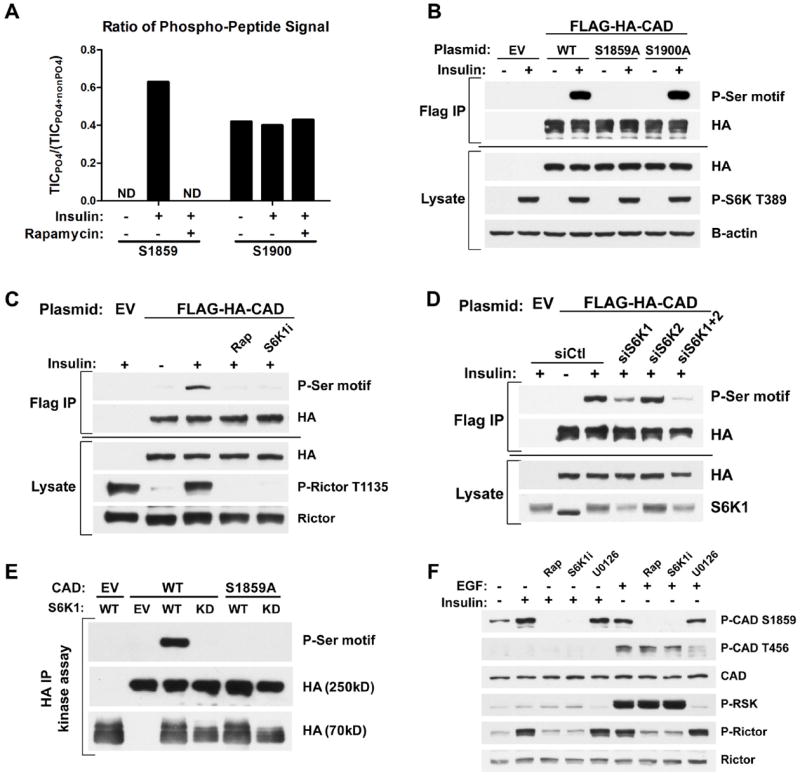
(A) Effects of insulin and rapamycin on CAD phosphorylation sites. FLAG-HA-CAD was immunopurified from serum-starved (16 h) HEK-293E cells, treated for 1h with DMSO or rapamycin (20 nM), prior to stimulation with insulin (3 h, 50 nM). The ratios of phosphorylated to total peptide levels, measured as total ion current (TIC) by LC/MS/MS, of the indicated sites on CAD under the different conditions are graphed. ND=phospho-peptide not detected.
(B) HEK-293E cells expressing empty vector (EV) or wild-type (WT), S1859A, or S1900A versions of FLAG-HA-CAD were serum-starved (16 h) and stimulated with insulin (1 h, 100 nM). FLAG-immunoprecipitates were immunoblotted with a phospho-14-3-3-binding motif antibody (P-Ser motif).
(C) Cells were treated as in (B), but pretreated for 1 h with rapamycin (20 nM) or the S6K1 inhibitor PF-4708671 (10 μM, S6K1i) prior to insulin stimulation.
(D) Cells were treated as in (C), but were also transfected with siRNAs targeting S6K1, S6K2, or both, or non-targeting controls (siCtl).
(E) In vitro kinase assays were performed with FLAG-HA-CAD substrate (WT or S1859A) immunoprecipitated from serum-starved, rapamycin-treated HEK-293E cells and HA-S6K1 (WT or kinase dead, KD) immunoprecipitated from insulin-stimulated HEK-293E cells.
(F) Hela cells were serum-starved (16 h) and pretreated for 1 h with rapamycin, S6K1i, or the MEK inhibitor U0126 (10 μM) prior to 1-h stimulation with insulin (100 nM) or EGF (20 ng/mL).
