Fig 4. Requirement of S6K1 and S1859 on CAD for the mTORC1-dependent stimulation of the de novo pyrimidine synthesis pathway.
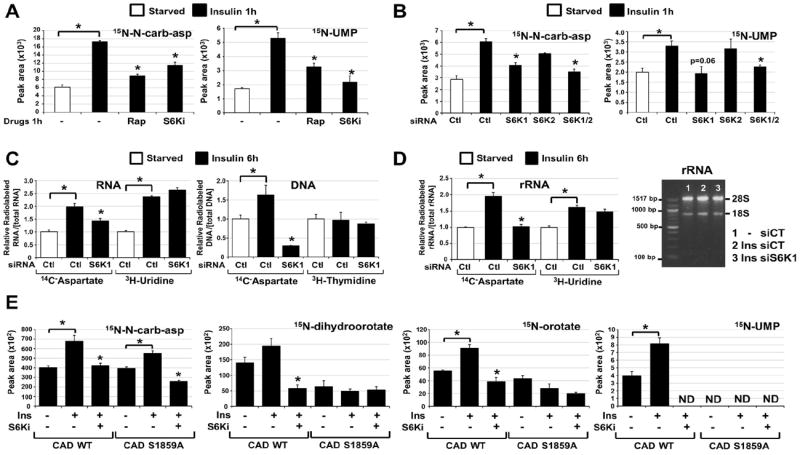
(A) Normalized peak areas of 15N-labeled metabolites, measured by LC/MS/MS, extracted from WT MEFs serum starved (15 h) and insulin stimulated (1h, 100 nM) in presence of DMSO, rapamycin (20 nM) or PF-4708671 (10 μM, S6Ki), with a 15-min pulse label of 15N-glutamine.
(B) Normalized peak areas of 15N-labeled metabolites from WT MEFs transfected with siRNAs targeting S6K1, S6K2, or both, or non-targeting controls (siCtl), were treated 48 h post-transfection as in (A).
(C) The relative incorporation of radiolabel from 14C-aspartate, 3H-uridine or 3H-thymidine into RNA and DNA from WT MEFs transfected with siRNAs as in (B), serum starved (15 h), and stimulated with insulin (6 h, 100 nM), during which cells were radiolabeled.
(D) The relative incorporation of radiolabel from 14C-aspartate or 3H-uridine into rRNA from WT MEFs treated as in (C). The purified rRNA was also assessed on an agarose gel (right).
(E) Normalized peak areas of 15N-labeled metabolites in G9C cells expressing CAD WT or a S1859A mutant treated as in (A). ND=metabolite not detected.
(A-E) All data are presented as mean±SEM over three independent samples per condition. *P<0.05 for pairwise comparisons calculated using a two-tailed Student’s t test (N=3), with all P-values provided in table S4.
