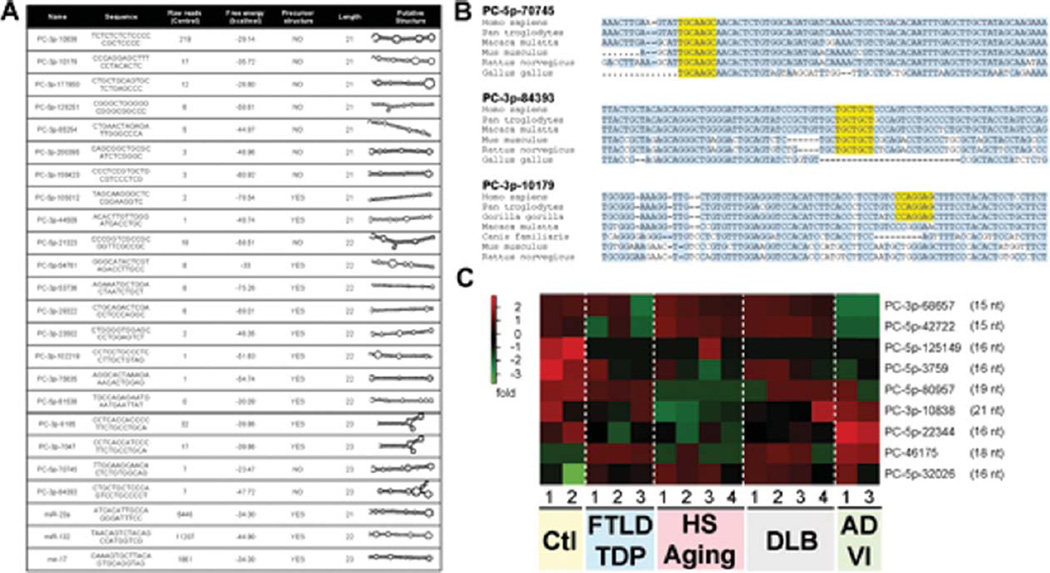
Fig. 5.
A) Table demonstrating putative novel miRNAs with 21–23 nucleotides. The candidate miRNA names (“PC-”), sequence, number of reads (average n = 2 of controls), precursor miRNA (pre-miR) free energy (calculated using RNAfold: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), miRNA precursor properties (calculated using MirEval: http://tagc.univ-mrs.fr/mireval), and putative pre-miR structure (generated using RNAfold). Range of read numbers in these samples was from 0 (PC-3p-81538) to 219 (PC-3p-10838). A “0” reads count indicates that no putative miRNA molecule was detected in this sample. For comparative reasons, we included miR-23a (21nt), miR-132-3p (22nt), and miR-17 (23nt) which all had >1000 reads. B) Conservation alignment of three putative novel miRNAs. The seed sequence (nucleotides 2–7 of the mature sequence) is shown in yellow. Conserved nucleotides are shown in light blue. C) Heatmap of significantly misregulated putative novel miRNAs in the different disease groups when compared to non-demented controls. Note that only one candidate miRNA has 21 nucleotides.