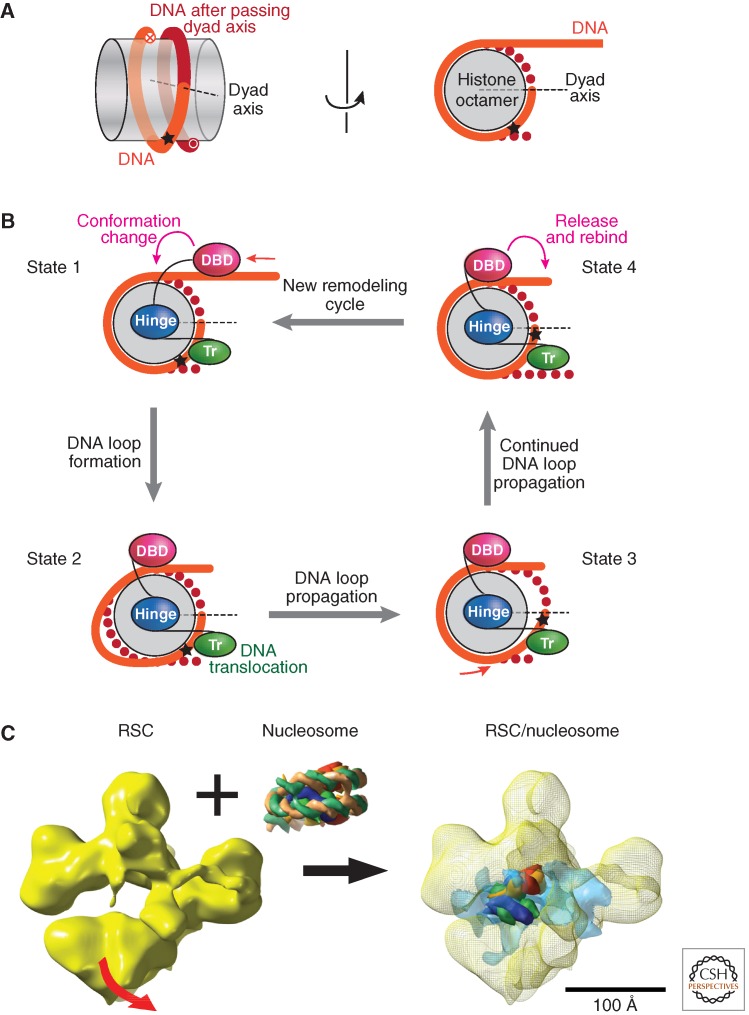
Figure 5.
Mechanism of nucleosome remodeling. (A) Nucleosome view emphasizing the left hand wrapping of DNA. (Left) Side view of the nucleosome core with the histone octamer represented as a gray transparent cylinder and the DNA in orange (before the dyad axis) and red (after the dyad axis). (Right) Top view of the nucleosome (rotated 90º) in which the DNA after the dyad axis is represented by red dots. The star represents a reference point on the DNA sequence. (B) Model for DNA movement across the histone octamer during a remodeling event by the ISWI-type enzymes. Successive steps in a remodeling event are represented by States I–IV. In State I the DNA binding domain (DBD) is bound to the linker DNA and the translocase (Tr) domain is bound near the nucleosome dyad. A hypothetical “hinge” mediates the changes in conformation. In State II a conformational change between the DBD and the Tr “pulls in” DNA, which becomes visible as a bulge on the histone octamer surface. The Tr activity propagates this bulge across the surface of the histone octamer beyond the dyad axis (State III). The DNA loop continues to diffuse across the octamer surface and is released into the distal linker DNA (State IV). Loop diffusion thus effectively repositions the histone octamer relative to the DNA sequence (i.e., the star has moved closer to the dyad axis). A further conformational change triggered by aspects of the ATPase cycle lead to a resetting of the remodeler relative to the histone octamer (compare in States IV and I). The remodeler now engages with a different segment of linker DNA to start another cycle of remodeling. (A,B, Adapted from Clapier and Cairns 2009.) (C) Cryo-electron microscopy (EM) analysis of the RSC structure and nucleosome interaction. The yeast RSC resembles the Swi/Snf complex in subunit composition and overall architecture. Instead of the ATPase Swi2/Snf2, it contains the Sth1ATPase, which belongs to the same subfamily (Clapier and Cairns 2009). A 25-Å cryo-EM map of RSC (left) shows a central cavity that closely matches the shape and dimensions of a nucleosome core particle. Movement (indicated by the red arrow) of the bottom RSC domain appears to control access to the central cavity. Incubation of RSC with nucleosome core particles (NCPs) results in formation of a RSC-NCP complex (right panel) in which NCP density is apparent in the central RSC cavity. Interestingly, interaction with RSC in the absence of any ATP hydrolysis appears to result in extensive changes in NCP organization. Histone density can be identified, but nucleosomal DNA appears disordered (semitransparent blue density). This loosening of DNA may facilitate DNA translocation during remodeling. (Image and interpretation provided by Francisco Asturias, Scripps Research Institute, La Jolla.)