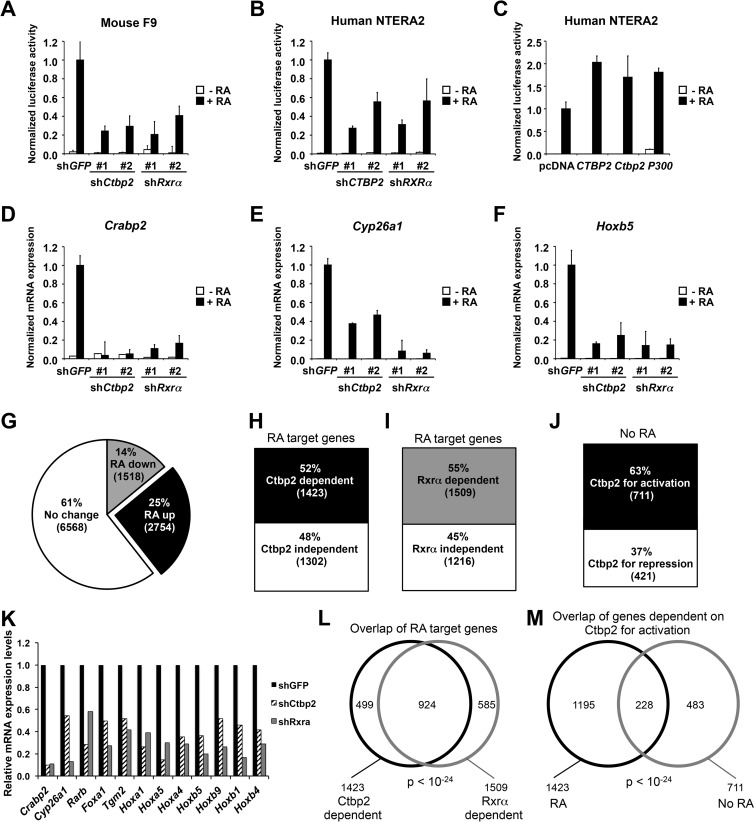
Fig 4.
CTBP2 is a transcriptional cofactor for RXRα/RARα. (A to C) Activation of a RARE-luciferase (RARE-Luc) reporter gene by RXRα/RARα requires CTBP2. The normalized luciferase activities shown represent ratios between luciferase values and Renilla internal control values and are the averages ± SD from three independent transfections. (A) Downregulation of Ctbp2 by RNAi inhibited the transcriptional induction of the RARE-Luc reporter gene in mouse F9 cells in response to 24 h of 1 μM RA stimulation. (B) Suppression of CTBP2 by RNAi inhibited the activation of the RARE-Luc reporter gene in human NTERA2 cells in response to 1 μM RA treatment. (C) Overexpression of human CTBP2, mouse Ctbp2, and human p300 histone acetyltransferase was able to hyperactivate the luciferase expression induced by 1 μM RA in the NTERA2 cell line. (D to F) Ctbp2 is required for transcriptional regulation of endogenous RA target genes in response to RA. Shown are the results from mRNA expression analysis of RA target genes Crabp2 (D), Cyp26a1 (E), and Hoxb5 (F) in F9 cells expressing shRNAs targeting Ctbp2, Rxrα, or GFP cultured in the absence or presence of 1 μM RA for 48 h. Error bars represent SD of triplicate independent experiments. (G) Pie chart showing the results from the RNA sequencing analysis in F9 shGFP control cells cultured in the absence or presence of RA for 48 h. Twenty-five percent (2,754) of the genes are upregulated in an RA-dependent manner, 14% (1,518) of the genes are downregulated in an RA-dependent manner, and 61% (6,568) of the genes remain unchanged. (H and I) Effect of Ctbp2 and Rxr knockdown on RA-upregulated genes by whole-genome transcriptome analysis in F9 cells. A bar chart shows that 52% of the upregulated genes are dependent on Ctbp2 (H) and 55% of the genes are dependent on Rxr (I) for transcription. (J) Ctbp2-dependent genes for activation and repression in the absence of RA (J). (K) Representation of the major bona fide RA target genes and their dependence on Ctbp2 and Rxrα for activation. (See Materials and Methods for details.) (L and M) Venn diagram representation of the overlap of genes between Ctbp2 and Rxr dependency (L). Shown is the overlap (228 genes) of Ctbp2-dependent genes for activation (711 genes) with the RA-dependent Ctbp2-dependent genes for activation (1,423 genes) (M).