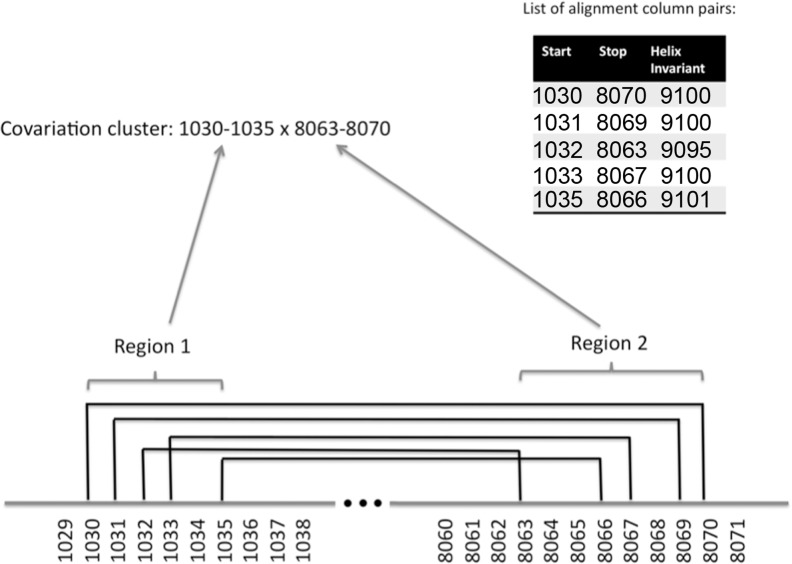
FIGURE 1.
Hypothetical example of a covariation cluster. Shown is a hypothetical chromosome with locations on that chromosome indicated by their nucleotide positions. A covariation cluster is a set of alignment column pairs with covariations that are grouped together by a clustering algorithm. Each alignment column pair with covariation is characterized by two genomic positions (called start and stop position) as well as the “helix invariant” (the sum of their respective start and stop positions—see Materials and Methods). In this hypothetical example, there are five column pairs with covariation (they are listed in the table embedded at the top-right of the figure; they are also depicted via black lines connecting the respective start and stop positions). Each covariation cluster consists of two genomic regions (called Region 1 and Region 2). Several conditions have to be fulfilled for a covariation cluster: (1) A covariation cluster has to contain at least five alignment column pairs with covariation; (2) the distance between the two genomic regions (in this example the distance between Region 1 and Region 2 is 8063–1035 = 7028) has to be at least 6000 nt; (3) the “difference” between the total number of covarying alignment column pairs of the cluster (in this example there are five alignment column pairs with covariation) and the number of “different” helix invariants (in this example there are three different helix invariants: 9095, 9100, 9101) has to be two units or larger; (4) the individual genomic regions have to have a minimum length of 5 nt (in this example the lengths of Regions 1 and 2 are 6 nt and 8 nt, respectively; in other words, in this example the covariation cluster would pass this particular filter criterion). The clustering algorithm (a single-linkage clustering) ensures that for every alignment column of a covarying alignment column pair there exists at least one other alignment column that belongs to another alignment column pair of the same cluster, such that their genomic positions differ by not more than 40 nt. Note that the two regions of a covariation cluster can be located on the same chromosome or on different chromosomes. Subsequent computational filtering stages are described in the subsection “Data processing steps” (Materials and Methods).