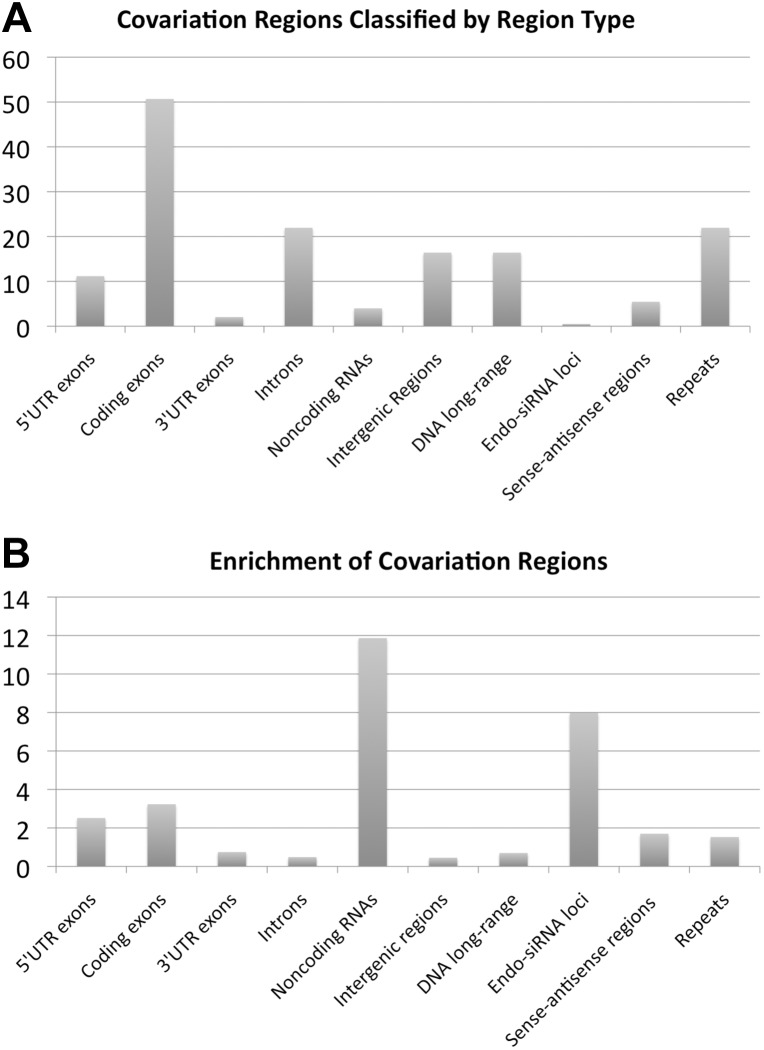
FIGURE 4.
Classification of genomic regions that are involved in long-range covariation. (A) Fraction (in percent) of covariation regions that overlap with genomic regions of a certain type. (B) Enrichment of a fraction of bases that are predicted to be within covariation regions. The enrichment of a region type is defined as (Ncr/Nr)/(Nc/N); Ncr: number of genome positions that overlap with at least one region of a predicted covariation cluster; Nr: number of different genome positions corresponding to the genomic region type of interest; Nc: total number of genome positions that overlap with at least one region of a predicted covariation cluster; N: total effective genome length. Note that the used uncorrected values of Ncr and Nc are affected by the restrictions w.r.t. the used available alignments that cover only 59% of the euchromatic genome. This bias toward the “alignome” is approximately canceled out in the above formula if the same correction factor of 0.59 is applied to both Nr and N. (Region types) DNA long-range: genomic regions that were previously reported to participate in long-range chromosomal interactions (Sexton et al. 2012); intergenic: all euchromatic genomic regions that are not annotated as coding exons, noncoding RNAs or introns; endo-siRNA loci: genomic regions reported to be involved in endo-siRNA generation (Czech et al. 2008). Note that because a genomic region can be annotated with several region-type attributes, the sum of the percentages do not have to add up to 100. The standard deviation (estimated using the standard deviation of a Poisson random variable combined with error propagation) relative to the magnitude of the computed enrichment is 4% in the case of the sense–antisense regions and below 0.01% for all other region types.