FIGURE 3.
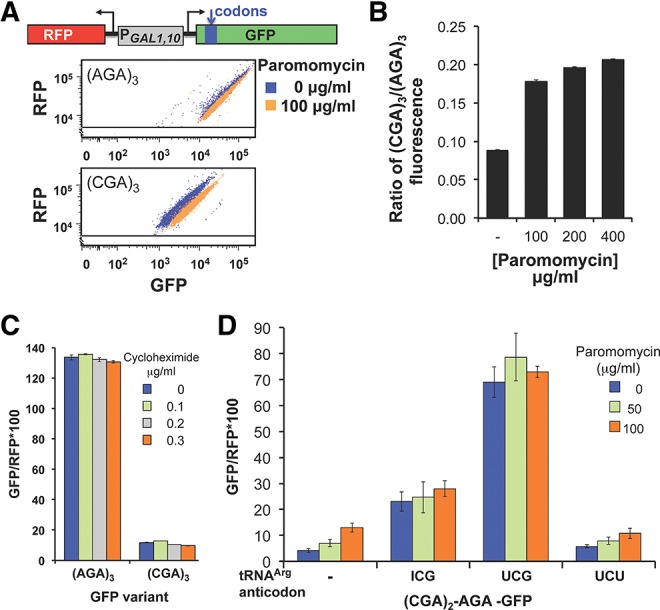
Paromomycin and increased amounts of native tRNAArg(ICG), but not cycloheximide, suppress expression of (CGA)3-GFP. (A) Addition of paromomycin results in increased GFP/RFP fluorescence of (CGA)3-GFP but not (AGA)3-GFP. Scatter plot of cells expressing (AGA)3-GFP or (CGA)3-GFP, grown with and without 100 μg/mL paromomycin. Schematic of the RNA-ID Arg4-GFP reporter used in A–D is included. The GFP variants each bear three codon insertions beginning at amino acid 4 or 6 (Dean and Grayhack 2012). (B) The expression of (CGA)3-GFP relative to that of (AGA)3-GFP increases mildly with increasing amounts of paromomycin. The average GFP/RFP is derived from the median GFP/RFP of at least three isolates of each construct. (C) Addition of cycloheximide has no effect on GFP/RFP fluorescence ratio of (CGA)3-GFP or (AGA)3-GFP variants. (D) Expression of either the native decoding tRNAArg(ICG) or the anticodon mutated tRNAArg(UCG) nearly abolish the effects of paromomycin. GFP/RFP fluorescence ratio of the [(CGA)2(AGA)1]-GFP variant as a function of increasing amounts of paromomycin is shown in strains bearing the indicated tRNAs on multicopy plasmids. tRNAArg species are indicated by the anticodon of the tRNA; the tRNA with the UCG anticodon was obtained by mutating the anticodon of tRNAArg(ICG) to UCG (Letzring et al. 2010).
