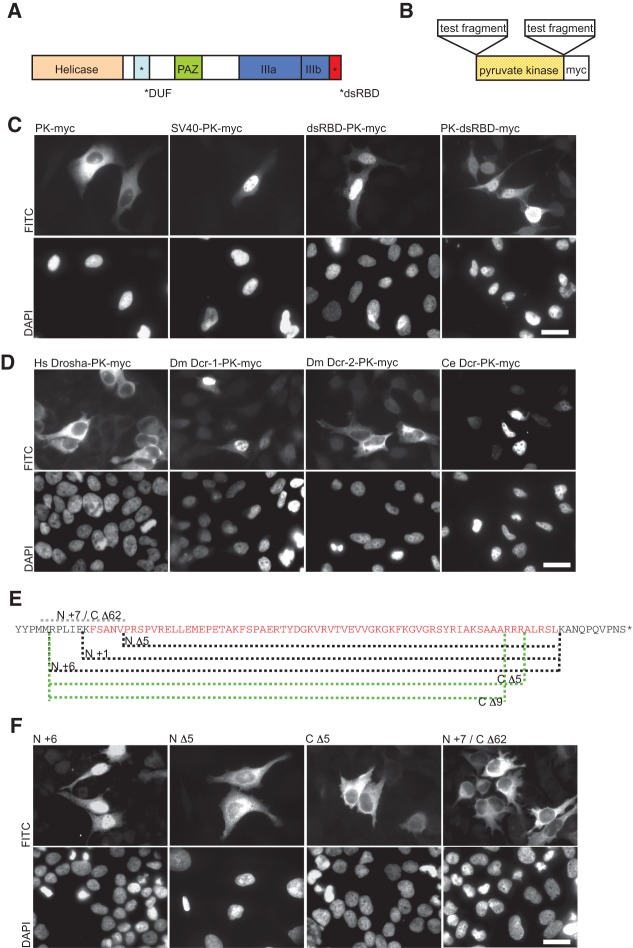
FIGURE 1.
The dsRBD of human Dicer functions as a nuclear localization signal (NLS). (A) Schematic representation of human Dicer with individual protein domains indicated by different colors. (B) The pyruvate kinase (PK-myc) fusion constructs used to assess NLS activity. (C) In transfected HeLa cells, PK-myc localizes to the cytoplasm but can be driven to the nucleus by an SV40 NLS (SV40-PK-myc). dsRBD-PK-myc and PK-dsRBD-myc show strong nuclear accumulation. Staining with DAPI is shown in the lower panel. (D) dsRBDs from various proteins were tested for their ability to function as a NLS in the PK reporter assay as indicated. The dsRBDs from human Drosha (Hs Drosha-PK-myc) and Drosophila Dcr-2 (Dm Dcr-2-PK-myc) were found not to function as a NLS, whereas those of Drosophila Dcr-1 (Dm Dcr-1-PK-myc) and C. elegans Dcr (Ce Dcr-PK-myc) functioned as a NLS. Bar, 20 µm. (E) Sequence of Dicer’s dsRBD (amino acids 1845 to 1912, in red) and its flanking amino acids (in black). The indicated deletions made at either the N or C terminus of the dsRBD are shown by dashed lines. The protein fragments of each dsRBD reporter tested are as follows: N Δ5, 1850 to 1912; N +1, 1844 to 1912; N +6, 1839 to 1912; C Δ5, 1839 to 1907; C Δ9, 1839 to 1903. N +7/C Δ62, including only residues 1837 to 1849, served as an additional control to test if residues spanning gaps between other deletion constructs can function as a NLS. (F) In transfected HeLa cells, Dicer dsRBD N +6 and N +1 (data not shown for N +1) accumulated in the nucleus. N- or C-terminal deletions (N Δ5, C Δ5, and C Δ9 [data not shown for C Δ9]) localize to the cytoplasm. The additional control N +7/C Δ62 also localizes to the cytoplasm. Staining with DAPI is shown in the lower panel. Representative images are shown from a minimum of three independent experiments. Bar, 20 µm.