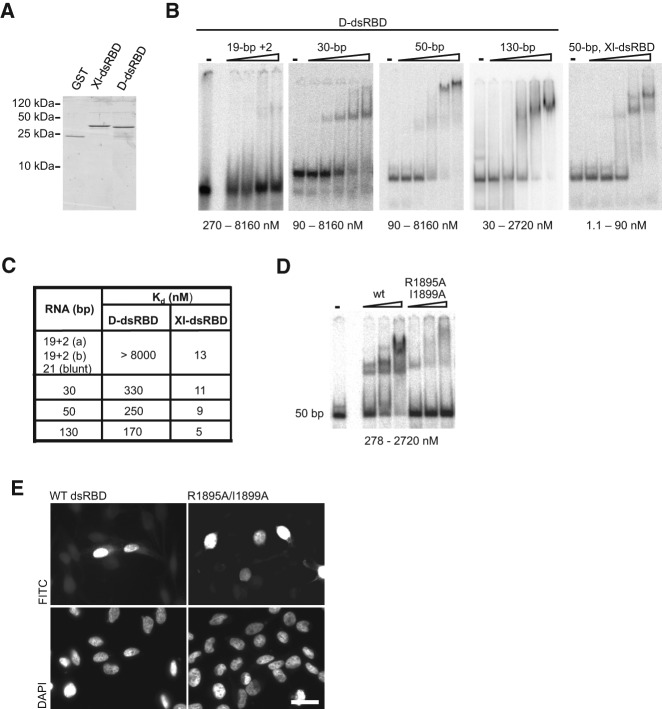
FIGURE 2.
RNA-binding potential of Dicer dsRBD is not required for NLS activity. (A) Dicer’s dsRBD (D-dsRBD) and the second dsRBD from the Xenopus laevis RNA-binding protein A (XlrbpA) (Xl-dsRBD) were expressed as GST-fusion proteins and purified from Escherichia coli. (B) To analyze the D-dsRBD RNA-binding properties, native gel mobility shift assays were performed three or more times with increasing amounts of recombinant D-dsRBD and substrates of differing length as indicated. D-dsRBD bound 30-, 50-, and 130-bp-long dsRNAs, but only little binding of the shorter 21- or 19-bp substrates was observed at protein concentrations tested. Two independent sequences for the 19-bp substrates containing 2-nt 3′-overhangs, 19 + 2 (a) and 19 + 2 (b), were tested. (C) Apparent binding affinities for the different substrates were measured for D-dsRBD and the Xl-dsRBD using the representative experiments shown in panel B. D-dsRBD bound longer dsRNAs with an affinity ∼30 times lower compared to the Xl-dsRBD. (D) Mutational analysis of D-dsRBD was performed (also see Supplemental Fig. S2) to identify residues important for RNA-binding. Introduction of the double point mutation (R1895A/I1899A) severely abrogated RNA-binding. (E) The NLS activity of the double point mutant (R1895A/I1899A) was tested in the PK-myc reporter assay and compared to the wild-type (WT) domain. Both fusion proteins efficiently accumulated in the nucleus. Staining with DAPI is shown in the lower panel. Representative images are shown from a minimum of three independent experiments. Bar, 20 µm.