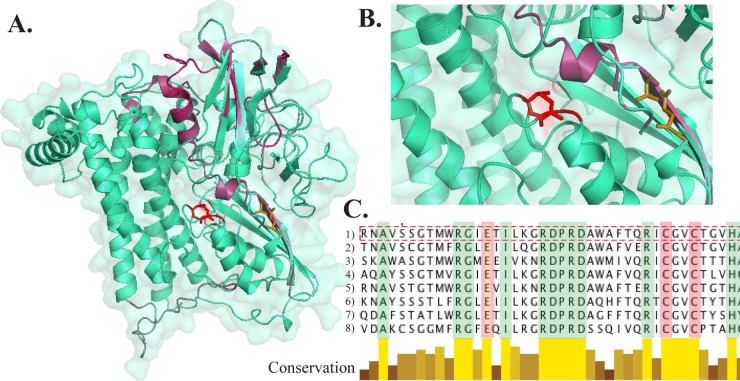
Fig 6.
(A and B) Three-dimensional (3D) structure representations of the hydrogenase HydB in Leptospirillum group IV modeled on the Escherichia coli protein. (A) HydB full model; (B) higher-magnification image of the active site highlighting catalytic residues (NiFe-binding residues [red] and Fe2-binding residues [brown]). Conserved structural regions are shown in blue-green, while less-conserved regions are shown in magenta. (C) Multiple-alignment screenshot showing catalytic residues (Cys75 and Cys78 [rose] and Glu56 [peach]) and other perfectly conserved residues around the active site (green). Alignment includes hydrogenase sequences from Leptospirillum group IV UBA BS (line 1), E. coli (lines 2 and 3), Allocromatium vinosum (line 4), Bradyrhizobium japonicum (line 5), Desulfovibrio vulgaris (line 6), Wolinella succinogenes (line 7), and Desulfovibrio baculatus (line 8). The shades of yellow and brown indicate degrees of conservation.