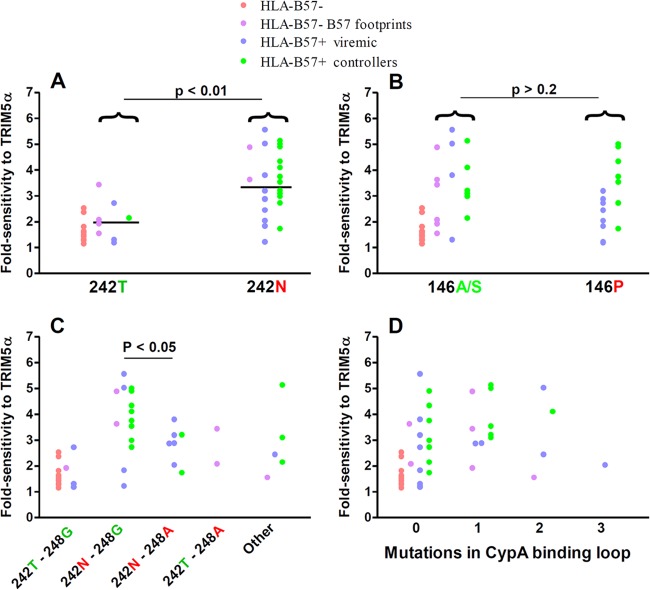
Fig 4.
Viral sensitivity to hTRIM5α and the CA genotype. Viral fold sensitivity to hTRIM5α was measured as described in the Fig. 1 legend and is shown for viruses expressing the indicated genotype at Gag amino acid 242 (A), viruses expressing the indicated genotype at Gag amino acid 146 (B), viruses expressing the indicated genotypes at Gag amino acids 242 and 248 (C), and viruses expressing the indicated number of compensatory mutations [H219Q, I223V, M228(I/L)] in the cyclophilin A binding loop (D). Symbols are color coded according to the patient groups indicated in the legend and described in the Fig. 1 legend. Bars indicate medians. Statistical analysis was performed using the Mann-Whitney test (panels A and B) or analysis of variance followed by Bonferroni's multiple comparison test (panels C and D). Note that these analyses were directed at identifying mutations selected by CTL targeting HLA-B57-restricted epitopes that modify hTRIM5α sensitivity. Consequently, results for HLA-B57− patients whose viruses did not express B57 footprint mutations (red dots) were excluded from the statistical analyses.