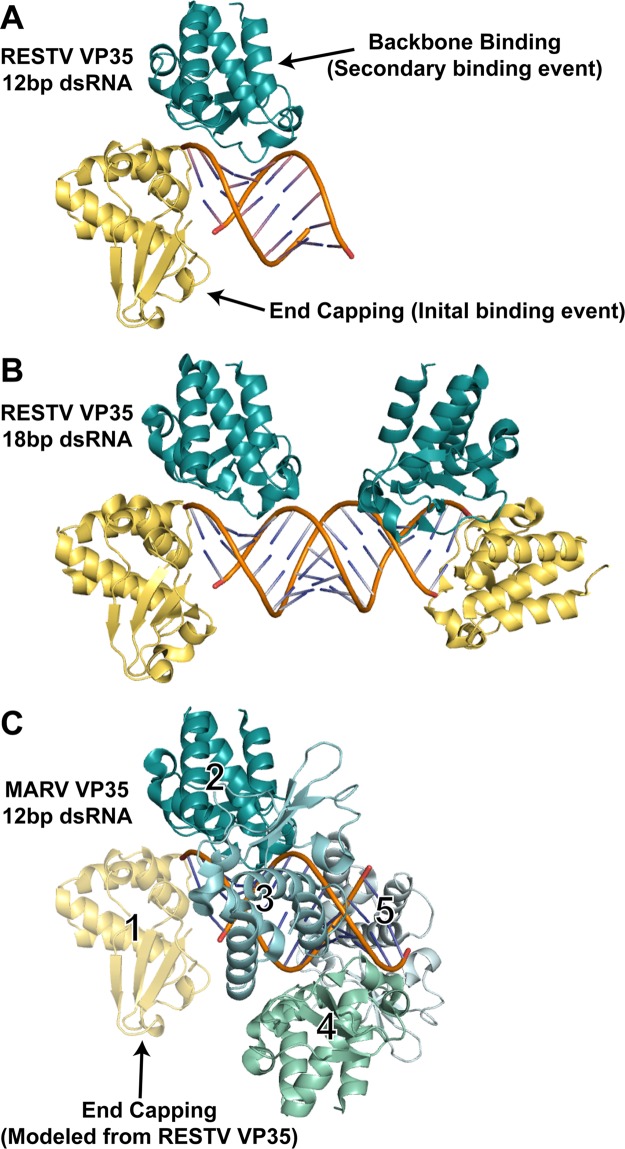
Fig 2.
dsRNA recognition events observed in different filovirus VP35-dsRNA complexes. (A) Crystal structure of the RESTV VP35 RBD in complex with a 12-bp dsRNA. The end-capping monomer (number 1 in panel C) is shown in yellow, and the backbone-binding member of the dimer (number 2 in panel C) is shown in teal. (B) Previous crystal structure of the RESTV VP35 RBD in complex with an 18-bp dsRNA (PDB accession no. 3KS8). (C) Continuous-coating crystal structure of the MARV VP35 RBD determined in complex with a 12-bp dsRNA (PDB accession no. 4GHA). Only backbone binding interactions are observed in the crystal structure (numbered 2 to 5, different shades of blue and green). An end-capping interaction (yellow), modeled from RESTV structures by the superimposition of the dsRNA and backbone-binding copy number 2 indicates that no steric clash would occur if MARV also bound an RBD on the dsRNA end (1).