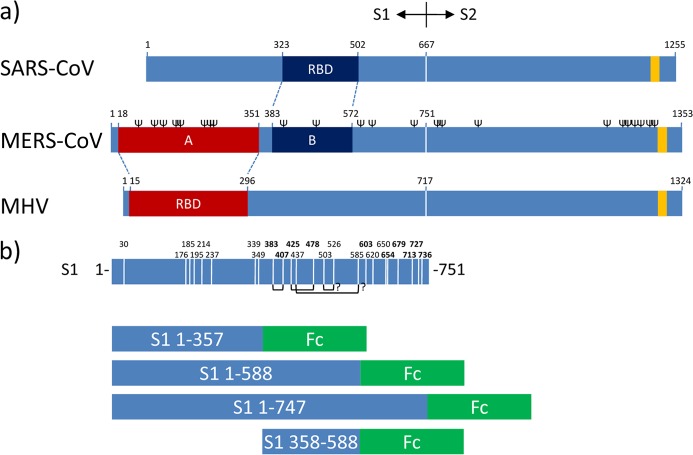
Fig 1.
RBDs in betacoronavirus spike proteins and S1-Fc expression constructs. (a) Schematic representation of the betacoronavirus SARS-CoV, MERS-CoV, and MHV (strain A59) spike (S) protein sequences (drawn to scale) aligned at the S1-S2 junction. The known RBD in the S1 subunit of the MHV and SARS-CoV S proteins and their corresponding homologous regions in MERS-CoV S as defined by ClustalW alignment are indicated. The positions of the transmembrane domains (yellow bars; predicted by the TMHMM server) and of the predicted N-glycosylation sites (Ψ; predicted by the NetNGlyc server, shown only for MERS-CoV S) are indicated. The border between the S1 and S2 subunits of the S protein is represented by a vertical white line. (b, top) Schematic representation of the MERS-CoV S1 subunit (residues 1 to 751) sequence. Cysteine positions in the S1 subunit are indicated by vertical white lines with the corresponding amino acid positions at the top. The positions of cysteines highly conserved among betacoronavirus S1 proteins are in bold. The predicted disulfide bond connections inferred from the structure of the SARS-CoV RBD are represented as connecting black lines at the bottom. (b, bottom) Domains of the MERS-CoV S1 subunit expressed as Fc chimeras.