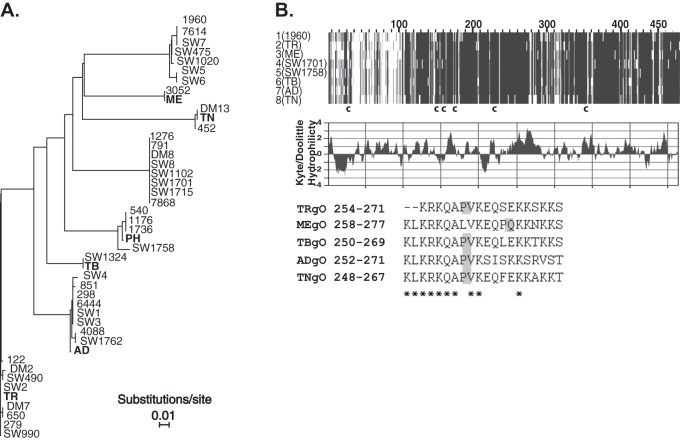
Fig 1.
Comparison of amino acid sequences of gO encoded by laboratory strains and clinical isolates of HCMV. (A) Phylogenetic relationships of predicted gO amino acid sequences of clinical HCMV isolates described in Rasmussen et al. (14) and the HCMV BAC clones used in the current studies (bold). Phylogeny was estimated using neighbor-joining with the Jones-Taylor-Thornton codon model implemented in MAFFT, version 7. Horizontal bars represent the expected number of amino acid substitutions per site according to the indicated scale. (B) MAFFT alignment of one representative gO sequence from each of the eight groups indicated in panel A is shown (top). Darker shading indicates sequence conservation. The approximate locations of six conserved cysteine (c) residues are indicated. The middle panel shows Kyte-Doolittle hydrophilicity analysis of TRgO. In the bottom panel are sequences of the synthetic peptides used to raise anti-gO antibodies. Asterisks indicate conserved residues, and shaded boxes highlight the position of a proline in each peptide sequence.