Fig 1.
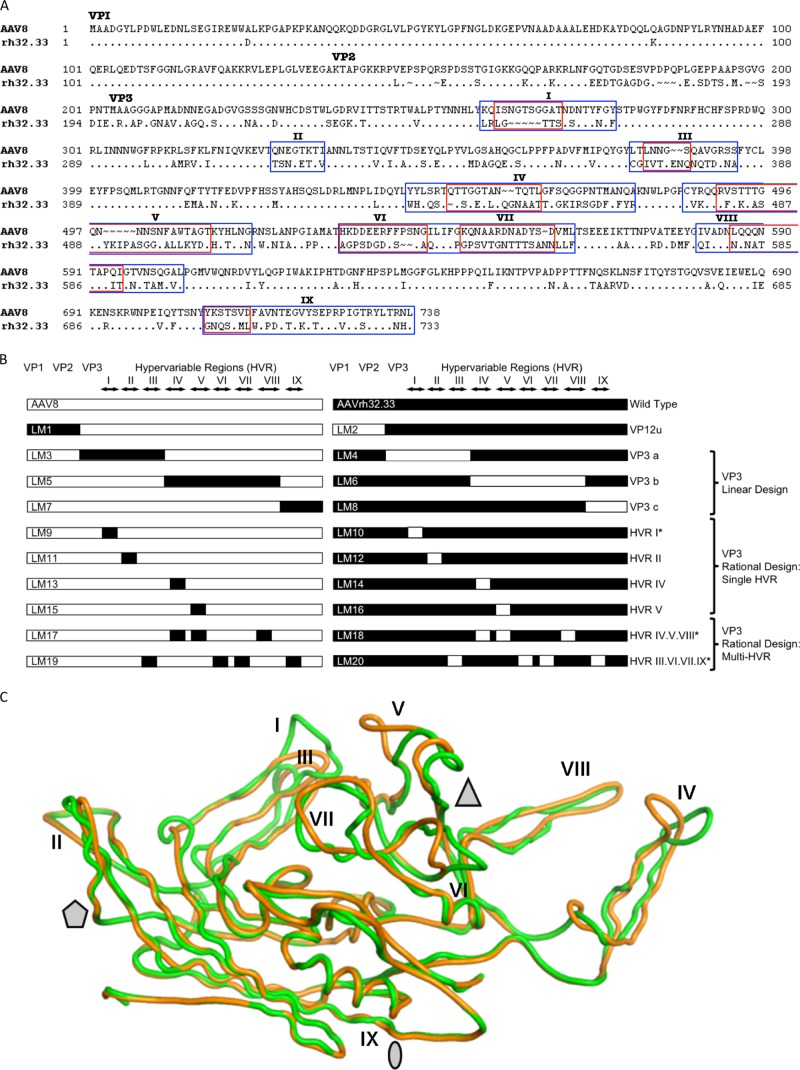
Comparison of the AAV8 and AAVrh32.33 capsids and the AAV8-AAVrh32.33 hybrid capsid constructs created by domain swapping. (A) Amino acid sequences of the VP1 capsid protein of AAV8 and AAVrh32.33 were aligned for comparison using ClustalX and BioEdit, version 7.0.0 (Ibis Biosciences, Carlsbad, CA). The dots in the alignment represent the amino acids that are identical to those of AAV8 VP1. The ∼ symbols indicate the amino acids that are missing at the positions in the alignment. The N termini for VP1 (ATG at M1), VP2 (ACG at T138), and VP3 (ATG at M204) are indicated. HVRs between β strands are numbered I to IX and enclosed by rectangular frames. Blue boxes, broad-swap delineations for each HVR within VP3; red boxes, narrow-swap locations for each HVR within VP3. Broad swaps contain all nonconserved amino acids within the given region. Narrow swaps contain only those amino acids that are structurally nonsuperimposable (defined as C-α atoms that are >1.0 Å apart); these are typically the most surface-exposed portions of each HVR loop. (B) Diagram of hybrid capsids constructed. Open bars, sequences from AAV8; closed bars, sequences from AAVrh32.33. The N termini of VP1, VP2, and VP3, as well as the delineations of each HVR, I to IX, are indicated at the top. Hybrid names are listed as abbreviations LM1 to LM20, with odd numbers representing hybrids composed primarily on an AAV8 backbone and even numbers representing hybrids composed on the AAVrh32.33 backbone. The swapped portion of the capsid as well as the design strategy for each set of hybrids is indicated to the right. *, hybrids that were constructed in both broad and narrow formats. (C) Superimposition of the VP3 monomers of AAV8 (green) and AAVrh32.33 (orange). Stretches of two or more C-α positions greater than 1 Å apart between the two structures were identified as variable loop regions. The major differences between AAV8 and AAVrh32.33 are located in HVRs I, IV, and V. When a second symmetry-related monomer interacts, HVR V falls in the groove between HVR VIII and HVR IV, shown in panel C to create the peaks surrounding the 3-fold axis of symmetry. The 2-fold (oval), 3-fold (triangle), and 5-fold (pentagon) axes of symmetry are indicated. In the LM14 hybrid, the inward-facing orange loop of AAVrh32.33 HVR IV is replaced with that of AAV8 (in green), opening up the gap between HVR IV and HVR VIII and exposing amino acid residues located within that groove.