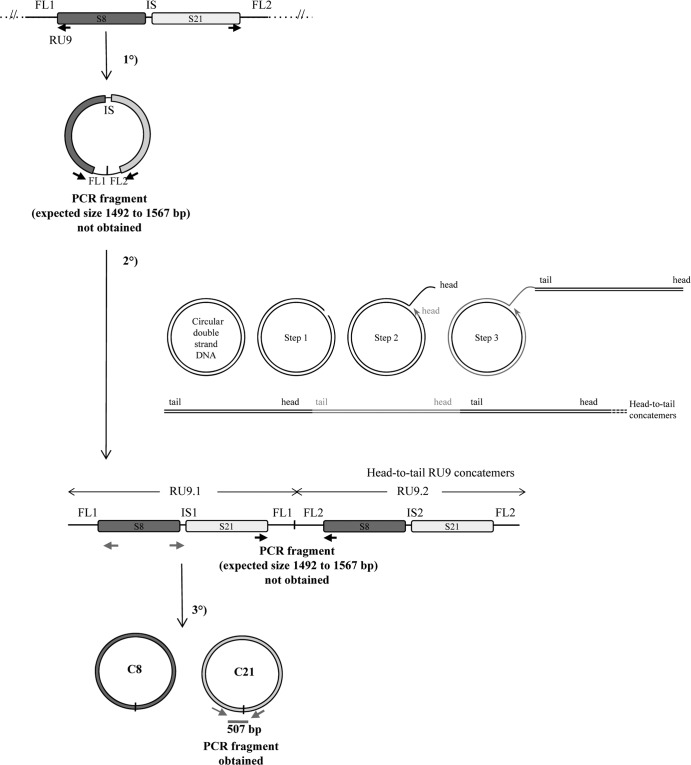
Fig 6.
Schematic representation of the macrocircle excision and rolling-circle amplification model (tested on RU9). The first step consists of the excision of a large circle from the wasp chromosome, including S8 and S21 sequences and their ANES (FL1-IS-FL2). This molecule is then used as a template for rolling-circle replication. After a nick on one strand of the circular dsDNA molecule (step 1), the 3′ OH end is released to serve as a primer for DNA synthesis by DNA polymerase (step 2 and step 3). Using the other strand as a template, replication proceeds around the circular DNA molecule, displacing the nicked strand as single-stranded DNA (step 3). The molecule produced is a head-to-tail concatemer, which can later be converted to double-stranded circles (C8 and C21 for RU9). This model predicts the production of a junction between ANES flanking sequences (FL1 and FL2) on the template, which is amplified in the concatemer and can be detected by a specific PCR, thus giving rise to an amplified fragment from 1,492 to 1,567 bp long, depending on the actual size of RU9 FL1 and FL2 (Table 1). No PCR fragment was obtained using primers in opposite orientations at the extremities of S8 and S21 (black arrows), while a product of the expected size (507 bp) was obtained using primers (gray arrows) in opposite orientations at the extremities of S21 (assessing the production of the C21 circle junction). These results indicated that the DNA used can be amplified by PCR but that the large circle and concatemers predicted by the model are most probably not produced (unless a particular structure prevents their amplification by PCR) and that bracovirus packaged-sequence amplification is achieved using a different mechanism.