Abstract
Aspergillus oryzae has been used in the food and beverage industry for centuries, and industrial strains have been produced by multiple rounds of selection. Targeted gene deletion technology is particularly useful for strain improvement in such strains, particularly when they do not have a well-characterized meiotic cycle. Phenotypes of an Aspergillus nidulans strain null for the CreB deubiquitinating enzyme include effects on growth and repression, including increased activity levels of various enzymes. We show that Aspergillus oryzae contains a functional homologue of the CreB deubiquitinating enzyme and that a null strain shows increased activity levels of industrially important secreted enzymes, including cellulases, xylanases, amylases, and proteases, as well as alleviated inhibition of spore germination on glucose medium. Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that the increased levels of enzyme activity in both Aspergillus nidulans and Aspergillus oryzae are mirrored at the transcript level, indicating transcriptional regulation. We report that Aspergillus oryzae DAR3699, originally isolated from soy fermentation, has a similar phenotype to that of a creB deletion mutant of the RIB40 strain, and it contains a mutation in the creB gene. Collectively, the results for Aspergillus oryzae, Aspergillus nidulans, Trichoderma reesei, and Penicillium decumbens show that deletion of creB may be broadly useful in diverse fungi for increasing production of a variety of enzymes.
INTRODUCTION
Aspergillus oryzae is a multicellular fungus that has been used for centuries for the production of Asian foods and beverages, including sake (rice wine) and shoyu (fermented soybean). Today it is also used industrially as a source of secreted enzymes, including cellulases, amylases, proteases, β-galactosidase, and lipase, and as a host for the production of heterologous proteins (1, 2).
creB was identified in the model filamentous fungus Aspergillus nidulans in a screen for mutations that alleviate carbon catabolite repression (CCR) (3). CCR is a mechanism by which genes for the utilization of nonpreferred carbon sources are repressed in the presence of preferred carbon sources; organisms thus avoid wasting energy producing enzymes for the degradation of complex carbon sources when more readily metabolized carbon sources are available. A. nidulans creB mutants grow well and alleviate CCR of various enzymes, including acetyl-coenzyme A (acetyl-CoA) synthetase, isocitrate lyase, acetamidase, and alcohol dehydrogenase (3, 4). In addition, A. nidulans creB mutants show a pleiotropic range of phenotypes apparently unrelated to CCR, including slightly reduced conidiation, reduced growth with quinate, proline, or glucuronate as the sole carbon source, enhanced growth on acetamide or acrylamide, resistance to molybdate, hypersensitivity to acriflavine, and reduced acidification of liquid growth media (3–5). creB encodes a deubiquitinating enzyme (DUB); DUBs are cysteine proteases that specifically cleave ubiquitin from ubiquitin-conjugated protein substrates (6, 7).
Other genes identified in the same screen for mutations that reduce CCR were creA and creC (3, 4). Molecular analysis of creA showed that it encodes a DNA-binding regulatory protein that is the major regulatory protein involved in CCR in A. nidulans (8) and many other multicellular fungi subsequently analyzed. creA mutations lead to derepression of a wider range of enzymes than creB mutations, but lack-of-function alleles lead to severe morphological effects (9, 10). Molecular analysis of creC revealed that it encodes a WD40-containing protein (11). The pleiotropic phenotypes of mutations in creB and creC are similar and nonadditive, and overexpression of creB can suppress the absence of creC, but not vice versa, indicating that CreB is the active partner (7).
The finding that creB mutations in A. nidulans reduce CCR without causing the severe morphological effects associated with mutations in creA makes creB an attractive target for mutagenesis in industrially useful strains. To date, creB disruption has been reported in two industrial sources of cellulase enzymes: Trichoderma reesei and Penicillium decumbens (12, 13). The T. reesei creB disruptant has increased growth on maltose, increased secretion of proteases, and greatly increased total secreted cellulase and xylanase activities in the absence of glucose. The P. decumbens creB deletion mutant has increased cellulase and xylanase activities and increased total secreted protein levels.
We deleted the creB gene from A. oryzae and analyzed the phenotypic effects, with an emphasis on the expression and secretion of industrially relevant enzymes, and we found that a null strain showed increased activity levels. Reverse transcription-quantitative PCR (RT-qPCR) experiments showed that this increase was due to increased levels of mRNA, indicating effects at the level of transcriptional control.
MATERIALS AND METHODS
Strains and media.
A. oryzae RIB40 (ATCC 42149) and NBRC 30105 (JCM02239) were obtained from the NITE Biological Resource Centre (NBRC), Japan. A. oryzae DAR3699, isolated during soy fermentation, was obtained from CSIRO Division of Food Science & Technology, Australia (14). The A. nidulans “wild-type” strain had the full genotype biA1; riboB2; niiA4, and the A. nidulans creB1937 strain had the full genotype yA1 pabaA1; creB1937; riboB2 (6).
Specific growth tests were undertaken in minimal medium (containing 0.05% KCl, 0.05% MgSO4, and 0.15% KH2PO4 plus traces of Na2B4O7, CuSO4, FePO4, MnSO4, NaMoO4, and ZnSO4, with the pH adjusted to 6.5) supplemented with the appropriate carbon and nitrogen sources, as indicated in each experiment. Unless otherwise specified, the nitrogen source was filter-sterilized 10 mM urea added after autoclaving; nitrate was not used because creB deletion reduces nitrate utilization. Solid medium also contained 1% agar.
Spore germination assays.
Spores were scraped from plates containing 1% sucrose after growth for 8 days at 30°C, vortexed in 0.1% Tween 20 to dislodge any hyphae, and centrifuged at 1,000 × g for 1 min. Pelleted spores were resuspended in fresh 0.1% Tween 20, vortexed vigorously to separate clumps, and counted with a hemocytometer. Plates were inoculated with 200 μl 0.1% Tween 20 containing 500 or 5,000 spores and incubated at 30°C.
Enzyme assays.
Spores were scraped from spread plates containing 1% sucrose supplemented with 0.1 M KCl to promote extensive sporulation (15). Spores were prepared as described above, except with centrifugation at 1,500 × g for 10 min. A suspension containing 2 × 107 spores per ml was prepared in 0.1% Tween 20, and 0.5 ml of this suspension was added to 50 ml autoclaved medium in a 250-ml Erlenmeyer flask, producing a final spore concentration of 2 × 105 spores per ml. Where necessary, xylan from oat-spelt (Fluka) was dissolved before autoclaving via extended heating on a hot plate with vigorous stirring. The starch used was analytical-grade soluble starch (Univar). Inoculated flasks were incubated at 30°C with shaking at 150 rpm. Because light has been shown to influence secreted cellulase levels in some fungi (16, 17), a glass-front incubator was used, and ordinary fluorescent indoor lighting was left on throughout incubation to maintain approximately constant ambient light. Under these conditions, all strains grew as discrete pellets. After 48 h, supernatant samples were added to a one-sixth volume of Complete protease inhibitor (Roche) and frozen at −80°C until analysis. Biomass samples were vacuum filtered through 55-mm filter paper circles (Whatman) that had been preweighed after drying at 65°C. Filters with biomass were then washed with 200 ml reverse osmosis (RO)-purified water, dried at 65°C to constant mass, and weighed. Total secreted cellulase, xylanase, and amylase activities were measured using EnzChek cellulase, xylanase, and amylase substrates (Invitrogen).
Quantitative real-time PCR.
For RNA preparation, conidial suspensions were prepared from 2-day-old conidia in 0.01% Tween 20, and 4.0 × 107 spores were added to 200 ml liquid medium, producing a final spore concentration of 2 × 105 spores per ml, in 1-liter flasks. The cultures were incubated overnight at 30°C with shaking at 150 rpm. Total RNA was isolated from mycelia grown under the specified conditions, using an RNeasy Plant minikit (Qiagen) according to the manufacturer's instructions. For cDNA synthesis, total RNA was treated with DNase (Promega). cDNA first-strand synthesis was performed using a Moloney murine leukemia virus (M-MLV) reverse transcriptase kit (Promega). The design of primers and the calculation of optimum annealing temperatures for PCR were performed using NetPrimer (www.premierbiosoft.com/netprimer/). RT-qPCRs were performed according to the instructions of Applied Biosystems. All experiments were performed with SYBR green as the detector, using an ABI Prism 7000 sequence detection system with a 2-step PCR and 60°C as the annealing temperature, unless otherwise stated.
The primers used for this study are shown in Tables 1 and 2.
Table 1.
Oligonucleotide primers used for RT-qPCR
| Organism | Primer | Primer sequence (5′ → 3′) | Exons whose boundary was crossed | Amplicon size (bp) |
|---|---|---|---|---|
| A. nidulans | alcA F | GAGGCTCTGGACTTCTTCGCT | 2 and 3 | 107 |
| alcA R | GCGATTCTGCCTTGTTCCATA | 2 and 3 | 107 | |
| tubC F | TAACCTGCTCAACCCTGTTCC | 5 and 6 | 137 | |
| tubC R | CATAGAGCACAGAGCAGTTTGGAC | 5 and 6 | 137 | |
| A. oryzae | β-tubulin F | GGTAACCAAATAGGTGCCGC | 4 and 5 | 80 |
| β-tubulin R | GAGGAGCCATTGTAAACACCG | 4 and 5 | 80 | |
| amyABC F | AGGCGTGTACTGTATCGGCG | 6 and 7 | 117 | |
| amyABC R | CGTTGAGGAGTGGATAGTAAATGG | 6 and 7 | 117 | |
| glaA F | AGGCAATCTTGAATAATATCGGC | 1 and 2 | 113 | |
| glaA R | CACGGGTCCAGGTATAGAAATAATG | 1 and 2 | 113 |
Table 2.
General oligonucleotide primers
| Primer | Sequence (5′ → 3′) | Purpose |
|---|---|---|
| creB_US_F | CGTTCGCTCTCTAACTCCGTC | Amplification of upstream region of creB for deletion construct |
| creB_US_R | CCCCATAATTGTCACAAC | Amplification of upstream region of creB for deletion construct |
| creB_DS_F | GAGGGATCAGGAAGCGAG | Amplification of downstream region of creB for deletion construct |
| creB_DS_R | CCAGCTATGTGACCCAGG | Amplification of downstream region of creB for deletion construct |
| ptrA_F | GACGGGCAATTGATTACG | Amplification of ptrA for deletion construct |
| ptrA_R | CTATCATGGGGTGACGATG | Amplification of ptrA for deletion construct |
| creB_US_R_Fus | CGTATAGATCAGCGGCACCCCATAATTGTCACAAC | Assembly of creB deletion construct by fusion PCR |
| creB_DS_F_Fus | CTCATCGTCACCCCATGATAGGAGGGATCAGGAAGCGAG | Assembly of creB deletion construct by fusion PCR |
| Ao_creB_US_F2 | ACCGCCAATCCACACGTC | Confirmation of replacement of creB at upstream end (Fig. 1) |
| pPTR_for_creB_US2 | GATAGTGTTGGGGTCCATGC | Confirmation of replacement of creB at upstream end (Fig. 1) |
| Ao_DS_creBKOtest_F | TATGTAAATGGCTGTGTCCC | Confirmation of replacement of creB at downstream end (Fig. 1) |
| Ao_DS_creBKOtest_R | ACCGTTCCCAAAACCTG | Confirmation of replacement of creB at downstream end (Fig. 1) |
| Ao_β-tub_RT_F_bridge | TTTTGGGATGGAGAATTACG | Semiquantitative RT-PCR of creB mRNA levels with respect to β-tubulin levels in A. oryzae DAR3699 |
| Ao_β-tub_RT_R | CTTGAAGAGCTCCTGGATGG | Semiquantitative RT-PCR of creB mRNA levels with respect to β-tubulin levels in A. oryzae DAR3699 |
| Ao_creB_RT_F_bridge | ACCTGCTCTGCTATCTTCCG | Semiquantitative RT-PCR of creB mRNA levels with respect to β-tubulin levels in A. oryzae DAR3699 |
| Ao_creB_RT_R | GCGAAGTTTTGATAGCGAAG | Semiquantitative RT-PCR of creB mRNA levels with respect to β-tubulin levels in A. oryzae DAR3699 |
Statistical analysis.
All claims of statistical significance are based on two-tailed two-sample Student's t tests assuming unequal variance.
RESULTS
The A. oryzae genome contains orthologues of genes involved in carbon catabolite repression in A. nidulans.
Although many genes have been shown to be affected by CCR in A. oryzae (18), there has been little research into the genetics of CCR in this fungus. We used published amino acid sequences of A. nidulans proteins to identify putative orthologues in the A. oryzae genome. Clear orthologues of creA, creB, and creC were identified (Table 3). When sequences were aligned, the A. oryzae CreB protein showed 63% amino acid identity with CreB from A. nidulans and 42% amino acid identity with Cre2 from T. reesei, and there was 38% identity when all three proteins were aligned, with conservation strongest in the six DUB homology domains surrounding conserved cysteine, aspartic acid, and histidine residues required for catalytic activity. CreB in A. nidulans and Cre2 in T. reesei share 41% amino acid identity.
Table 3.
Conservation of proteins involved in CCR in A. nidulans and A. oryzaea
| Protein | % Identity | % Similarity |
|---|---|---|
| CreA | 84.7 | 87.1 |
| CreB | 73.5 | 78.5 |
| CreC | 80.1 | 83.9 |
| CreD | 81.7 | 85.8 |
| AcrB | 77.2 | 81.6 |
Putative A. oryzae orthologues of genes involved in CCR in A. nidulans were identified by BLAST searching of the A. oryzae genome, and putative amino acid sequences were determined. Orthologous amino acid sequences were aligned using Gap (GCG).
Creation of A. oryzae creB deletion and complemented strains.
A deletion construct was made which lacks 2,191 bp of creB, beginning from 4 nucleotides past the start codon, and contains 621 bp 5′ of creB and 769 bp 3′ of creB, surrounding the ptrA1 pyrithiamine resistance selectable marker (19). The plasmid was used to transform RIB40, and pyrithiamine-resistant colonies were obtained. To verify that a deletion of creB was obtained, one colony was analyzed using PCR to show that both ends of creB were replaced (Fig. 1). To confirm that the phenotype was due to the deletion, the strain was further transformed with a plasmid containing creB to produce a complemented strain for use as a control in experiments. To do this, a chlorate-resistant sector was isolated and shown to be a niaD mutant, as it was complemented by the A. nidulans niaD gene (20). This creBΔ;niaD1 mutant was transformed using a plasmid containing both A. oryzae creB and A. nidulans niaD, and colonies that grew on nitrate were selected and tested.
Fig 1.
Verification of gene replacement. (A) Schematic showing primer binding sites. Primers (see Table 2 for details): 1, Ao_creB_US_F2; 2, pPTR_for_creB_US2p; 3, Ao_DS_creBKOtest_F; 4, Ao_DS_creBKOtest_R. (B) Agarose gel showing PCR products. Lane 1, molecular size markers; lane 2, blank; lanes 3 to 6, primers and DNA templates for strains, as indicated. +, creB+ strain; Δ, creBΔ strain.
Growth of A. oryzae creBΔ strain.
Growth phenotypes of the RIB40 parent strain, the creB deletion strain, and the creB deletion strain complemented with creB were tested under a range of conditions (Fig. 2). The deletion mutant strain grew well on standard minimal media such as Czapek-Dox medium, which contains 3% sucrose as a carbon source, and on richer media such as potato dextrose agar (which contains starch and 2% glucose), albeit with slightly reduced conidiation and mycelial density. The strain also grew robustly in liquid culture: after 48 h, the biomasses of A. oryzae creBΔ triplicate spore-inoculated cultures grown in sorbitol, xylan, or xylan plus sucrose were not significantly different from the biomass of the wild-type or complemented strain (Fig. 3A).
Fig 2.

Phenotypic effects of deleting creB in A. oryzae. The indicated strains were grown for 4 days at 30°C on the indicated media. Sucrose was added at 1%; proline, acetamide, glucuronate, and quinate were added at 50 mM, as carbon sources; urea, proline, and acetamide were added at 10 mM, as nitrogen sources; and bovine serum albumin (BSA) was added at 0.01%. PDA, potato dextrose agar.
Fig 3.
Deletion of creB increases secretion of cellulases and xylanases under inducing conditions. Triplicate spore-inoculated 50-ml shake flask cultures of the three strains were grown under noninducing (1% sorbitol), inducing (1% xylan), and repressing (1% xylan plus 2% sucrose) conditions. After 48 h, biomass and supernatants were harvested, and total secreted enzyme activities were measured using EnzChek cellulase substrate or EnzChek xylanase substrate and are expressed as activities per unit of dry weight biomass, normalized such that activity in the wild-type strain under inducing conditions was 100 units. (A) Growth in liquid medium. (B) Total secreted cellulase activity. (C) Total secreted xylanase activity.
The deletion mutant exhibited pleiotropic phenotypes similar to those seen in A. nidulans, with commonalities including reduced growth on quinate, proline, or glucuronate as the sole carbon source, reduced growth on nitrate as the sole nitrogen source, enhanced growth on acetamide, resistance to molybdate, and hypersensitivity to high concentrations of acriflavine. In contrast to the case for the creB disruptant in T. reesei, improved growth was not observed on maltose, likely because secreted α-glucosidase activity would not limit growth of A. oryzae RIB40, which was selected for strong starch degradation (21).
Of particular note, the A. oryzae creBΔ strain showed greatly enhanced growth on the protein bovine serum albumin, with a slightly broader halo of degraded protein on plates containing milk. Together, these observations indicate increased protease secretion in the creB-deleted strain.
RIB40 and the creBΔ and complemented strains were germinated on glass coverslips in 1% sorbitol and 10 mM urea liquid medium and examined microscopically after 18 and 24 h. No differences in morphology, hyphal length, or the amount of branching between the strains were apparent. Similarly, when conidia of the three strains were inoculated on solid medium lacking either a carbon or nitrogen source, the spidery hyphal extensions had similar diameters and were present in similar amounts. When the three strains were grown with shaking in liquid medium for RNA and enzyme analyses (see Materials and Methods), all strains grew as discrete pellets, and thus there was no apparent effect on viscosity due to deletion of creB.
Deletion of creB alleviates glucose inhibition of conidial germination.
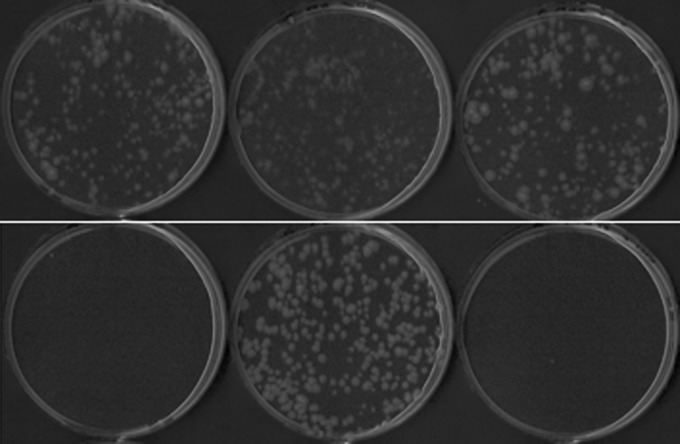
A. oryzae RIB40 germinates very poorly from conidial spore suspensions on 1% glucose medium and somewhat poorly on 1% sucrose medium compared to medium containing 1% fructose as the carbon source (Fig. 4; Table 4), indicating that glucose inhibits germination. This finding has not previously been reported explicitly, and the reason for it is not understood. This inhibition is not specific to the RIB40 strain, as it was also seen in A. oryzae NBRC 30105. Inhibition was not detected in the presence of a low concentration of glucose (0.1%) or on rich medium containing 1% glucose together with yeast extract, peptone, and amino acids (data not shown). The inhibition of conidial germination was abolished in the creBΔ strain (Fig. 4; Table 4).
Fig 4.
Inhibition of germination of A. oryzae by glucose. Strain RIB40, the creBΔ strain, and the creBΔ::creB+ strain (left to right) were grown from 500 spores for 2 days at 30°C on 1% fructose (top) or 1% glucose (bottom).
Table 4.
Deletion of creB alleviates inhibition of spore germination in the presence of repressing carbon sourcesa
| Carbon source | Spore germinationb |
||
|---|---|---|---|
| Wild-type strain | creBΔ strain | Complemented strain | |
| 1% fructose | +++ | +++ | +++ |
| 1% sucrose | ++ | +++ | ++ |
| 1% sucrose + 1% glucose | + | +++ | + |
| 1% glucose | —* | +++ | —* |
| 0.1% glucose | +++ | +++ | +++ |
Five hundred spores were spread on agar plates containing the indicated carbon sources as described in Materials and Methods.
Symbols indicate the numbers of colonies visible after 2 days of incubation. —, 0 colonies; +, 2 colonies; ++, 75 to 80 colonies; +++, >200 colonies. Similar trends were observed on plates inoculated with 5,000 spores (data not shown). * indicates colonies present at higher-density plating.
Deletion of creB increases expression of cellulases and xylanases under inducing conditions.
Preliminary experiments in RIB40 found that 1% sorbitol can be used as a carbon source that is neither an inducer nor a repressor of cellulases, amylases, and xylanases and that 2% sucrose causes repression of these enzymes. Subsequently, triplicate spore-inoculated 50-ml shake flask cultures of the three strains were grown under noninducing (1% sorbitol), inducing (1% xylan), and repressing (1% xylan plus 2% sucrose) conditions. After 48 h, total secreted cellulase activities were measured using EnzChek cellulase substrate and are expressed as activities per unit of dry weight biomass, normalized such that activity in the wild-type strain under inducing conditions was 100 units (Fig. 3B). Activity was barely detectable for all three strains under noninducing conditions, and the three strains' activities did not differ significantly. Under repressing conditions, activities were low but detectable and did not differ significantly between the three strains. Activities were over 2 orders of magnitude higher under inducing conditions. The wild-type and complemented strains' activities were not significantly different, whereas the creBΔ strain had significantly greater activity (P < 0.05), with the mean for the creBΔ strain cultures being 50% higher than that for the wild-type strain.
The same cultures were also analyzed for total secreted xylanase activity, using EnzChek xylanase substrate, and the results are expressed and normalized in the same manner as that described above (Fig. 3C). The results mirrored those for the cellulase assays, consistent with reports that cellulases and xylanases are regulated similarly in A. oryzae (22). Activities were barely detectable in all three strains under noninducing conditions, and the three strains' activities did not differ significantly. Under repressing conditions, activities were low but detectable and did not differ significantly between the three strains. Activities were over 3 orders of magnitude higher under inducing conditions. The wild-type and complemented strains' activities were not significantly different, whereas the creBΔ strain had significantly greater activity (P < 0.05), with the mean for the creBΔ strain cultures being almost double that for the wild-type strain.
Thus, deletion of creB increases the expression of cellulases and xylanases in the absence of CCR but does not affect the response to CCR.
Deletion of creB increases expression of amylases under various conditions via an increase in gene transcription.
Triplicate spore-inoculated 50-ml shake flask cultures of the three strains were grown under noninducing (1% sorbitol), inducing (1% sorbitol plus 1% starch), and repressing (1% sorbitol plus 1% starch plus 2% sucrose) conditions. After 48 h, total secreted amylase activities were measured using EnzChek amylase substrate and are expressed as activities per unit of dry weight biomass, normalized such that activity in the wild-type strain under inducing conditions was 100 units (Fig. 5A).
Fig 5.
Deletion of creB increases secretion of amylases under various conditions via an increase in gene transcription. (A) Triplicate spore-inoculated 50-ml shake flask cultures of the three strains were grown under noninducing (1% sorbitol), inducing (1% sorbitol plus 1% starch), and repressing (1% sorbitol plus 1% starch plus 2% sucrose) conditions. After 48 h, biomass and supernatants were harvested, and total secreted amylase activities were measured using EnzChek amylase substrate and are expressed as activities per unit of dry weight biomass, normalized such that activity in the wild-type strain under inducing conditions was 100 units. (B) qRT-PCR analysis of total α-amylase transcript levels in A. oryzae RIB40 and the creBΔ strain. (C) qRT-PCR analysis of glucoamylase A transcript levels in A. oryzae RIB40 and the creBΔ strain. Strains were induced using starch (1%) or repressed using sucrose (2%) and were grown at 30°C for 24 h. Transcript levels were standardized against β-tubulin levels. (D) qRT-PCR analysis of alcA transcript levels in A. nidulans wild-type and creB1936 strains. Strains were induced using EMK (50 mM) or repressed using glucose (1%) and were grown at 37°C for 16 to 18 h. Transcript levels were standardized against β-tubulin levels. Results shown are fold changes compared to the wild type induced with ethanol.
Under all three growth conditions, the activity of the creBΔ strain was significantly higher than that of the wild-type or complemented strain (P < 0.001 for noninducing conditions, P < 0.01 for inducing conditions, and P < 0.001 for repressing conditions). The activities of the wild-type and complemented strains were not significantly different from one another under any growth condition. The creBΔ strain produced readily detectable activity under noninducing conditions, with more than double the wild-type activity under inducing conditions and 40-fold more than the wild-type activity under repressing conditions.
The addition of 2% sucrose reduced amylase activities in the wild-type and complemented strains (P < 0.01) about 35-fold. It also reduced amylase activities in the mutant strain, but only by one-third. This indicates a high level of carbon catabolite derepression of one or more genes encoding starch-degrading enzymes in the mutant strain.
To investigate the molecular basis of these observations, glucoamylase (glaA) and α-amylase (amyA, amyB, and amyC) transcript levels in the wild-type and creBΔ strains were measured under all three growth conditions, using quantitative real-time PCR. Representative results from three independent experiments are shown in Fig. 5. There are three genes for secreted α-amylase in A. oryzae RIB40, with almost identical nucleotide sequences, which are all expressed (23, 24); our measurements indicate the total transcript levels of these three genes combined. The transcript levels of both glucoamylase (Fig. 5B) and α-amylase (Fig. 5C) were significantly higher (P < 0.01) in the creBΔ strain than in the wild-type strain under all three growth conditions, reflecting the higher secreted total amylase activities observed. Thus, creB deletion increases total secreted amylase activity by increasing transcript levels of multiple amylase-encoding genes.
The finding that deletion of creB in A. oryzae elevates the levels of glucoamylase and α-amylase transcripts and almost abolishes repression led us to look at an A. nidulans example, as the effects of creB on transcription have not been published. We chose alcA, encoding alcohol dehydrogenase I, as it has been well characterized at the plate test and enzyme activity levels and shows derepressed expression in medium containing both an inducer and a repressor (3). To investigate the molecular basis of this derepressed expression, transcript levels of the wild-type and creB1937 null strains grown under uninduced, ethyl methyl ketone (EMK)-induced, and EMK-plus-glucose-repressed conditions were measured using quantitative real-time PCR. Representative results from three independent experiments are shown in Fig. 5D. The data were assessed for the effects of creB1937 on the elevation of transcript levels, the inducibility of the system, and derepression. The transcript levels in the uninduced cultures showed that there was very low basal transcription, with no evidence of elevation of alcA transcription due to creB1937. The transcript levels increased significantly (P < 0.001) in the induced samples compared to the uninduced levels, with the increase being greater in the creB1937 strain than in the wild-type strain. While the strains induced with EMK showed very large elevations in transcription of alcA, there was a significant decrease (P < 0.001) when a repressor was added. In the wild type, this decrease was to the uninduced level; however, in the creB1937 strain, there remained, on average, a 20-fold increase in alcA transcript levels compared to that in the uninduced culture. These experiments were replicated using ethanol rather than EMK as the inducer of alcA, and the absolute levels of induction were reduced as expected, but the same results regarding elevation and repression were apparent. Thus, in A. nidulans, creB1937 leads to both elevation and partial derepression of alcA transcription.
A. oryzae DAR3699 has a phenotype similar to that of RIB40 creBΔ.
DAR3699, an A. oryzae strain from the CSIRO collection, was originally isolated during soy fermentation. Previous analyses found it to secrete high levels of amylase, to have a good growth rate, and to grow as compact pellets suitable for biomass production (14). As these properties make it suitable for bioreactor use, we included it in our phenotypic tests. DAR3699 showed phenotypes that were similar to those of the RIB40 creBΔ strain, including molybdate resistance, strong growth on acetamide, weak growth on proline, quinate, and arabinose, and high protease secretion. As indicated above, glucose inhibited the germination of RIB40 spores, but creB deletion abolished this inhibition; this inhibition was also absent in DAR3699. Furthermore, when we deleted creB in the DAR3699 strain, the phenotype was unchanged from that of DAR3699. These phenotypes indicated that DAR3699 might be a creB mutant strain and that this might contribute to its useful properties. We sequenced 8 kb of genomic DNA covering at least 200 bp upstream and 100 bp downstream of the creA, creB, and creC loci and found no differences between RIB40 and DAR3699 for creA and creC. The DAR3699 creB locus contains a single base pair insertion in a putative upstream open reading frame, lengthening it to 46 codons (Fig. 6). To test whether this change led to reduced transcription, RNAs were extracted from RIB40 and DAR3699 grown as surface colonies on liquid medium containing 1% glucose, and primers that amplify creB or β-tubulin were used in semiquantitative RT-PCRs. No reduction was detected in the amount of creB mRNA in DAR3699 compared with RIB40; thus, if the insertion in DAR3699 affects CreB levels, the effects are likely to be at the translational level. This suggestion is supported by findings in Saccharomyces cerevisiae which have shown that translation efficiency decreases markedly as upstream open reading frame length is increased, with 36 codons reducing translation by about 95% (25).
Fig 6.
Analysis of DAR3699 creB locus. The schematic shows a representation of the region at the 5′ end of the creB gene in A. nidulans, A. oryzae RIB40, and A. oryzae DAR3699 (not to scale). The top line represents the 5′ region of A. nidulans creB, showing the mapped start points of transcription (6) and an upstream open reading frame of 12 codons that is conserved in A. oryzae. The middle line represents the 5′ region of A. oryzae RIB40 creB, showing a putative 11-codon upstream open reading frame spanning the site that is the major start point of transcription in A. nidulans. The bottom line represents the 5′ region of A. oryzae DAR3699 creB, showing the effect of the insertion of one base pair into the upstream open reading frame sequence, lengthening it to 46 codons.
DISCUSSION
A. oryzae is used widely in the sake and soy brewing industries, as it produces and secretes a variety of amylases, cellulases, and proteases to break down carbohydrates and proteins in rice, wheat, and soybean to produce nitrogen and other nutrients, and these enzymes are also used to accelerate hydrolysis of substrates in fish sauce fermentation (1). Production strains have been selected over centuries for enhanced properties, but further improvements are possible.
Many of the genes encoding industrially important enzymes are not expressed when a good carbon source is available, due to the transcriptional control mechanism of CCR. For example, the gene expression profiles of A. oryzae cells were analyzed for mycelia grown in glucose-rich and glucose-poor media. A key finding was that cultures grown with wheat bran mimicked the glucose-depleted cultures and showed a diverse gene expression profile for hydrolytic enzymes, most probably due to a relaxation of CCR (18). Thus, an important approach to strain development is to perturb CCR in production strains, using information from model fungi and techniques that allow precise gene manipulations, to increase production of useful enzymes. Gene targeting has advantages over random mutagenesis, particularly in introducing multiple desired changes in organisms where genetic crosses cannot be undertaken due to the lack of a sexual cycle. Even though functional mating type genes were recently identified in A. oryzae (26), because production strains have a range of lesions in their genomes, molecular rather than meiotic approaches will still be preferable to maintain the overall phenotypes.
In A. nidulans, deletion of creA, encoding the major CCR repressor, has severe effects on morphology, so to avoid these effects, we chose to disrupt the A. oryzae creB gene. In A. nidulans, the regulatory deubiquitinating enzyme CreB is proposed to be involved in carbon metabolism by two possibly distinct mechanisms (3, 4). First, under carbon catabolite-repressing conditions, null alleles lead to derepressed expression of enzymes, including alcohol dehydrogenase I and acetamidase. Second, on a range of carbon sources generally not considered to be repressing, null alleles show altered growth, such as increased growth on maltose and decreased growth on d-quinate. Complete details of the mechanism of action of CreB are not fully understood, but the reduced growth on compounds such as quinate and proline is due to a direct targeting of permeases by CreB that is required to prevent premature turnover (27; N. Kamglangdee and J. M. Kelly, unpublished data). We demonstrate here that when A. oryzae lacks the CreB deubiquitinase, it produces higher levels of amylase, cellulase, xylanase, and protease activities. For cellulase and xylanase, these increased levels were apparent when no repressing carbon source had been added to the medium but not when a repressing carbon source was exogenously added, whereas amylase expression was increased under both conditions. Thus, we propose that A. oryzae CreB is involved not only in derepression of some enzymes but also in the expression of some enzymes even in the absence of CCR, consistent with observations in A. nidulans (3–6). The molecular basis of the creB deletion derepression phenotypes is not understood for any organism, so to determine whether there are effects on transcription, glucoamylase and α-amylase transcript levels were compared between the wild-type and creBΔ strains, and enzyme activity changes were found to be mirrored at the transcript level. This finding is significant, and because there are no published data about the transcriptional effects of creB mutations in A. nidulans, we examined alcA expression. alcA encodes alcohol dehydrogenase I, has been well characterized at the plate test and enzyme activity levels, and shows derepressed activity in medium containing both an inducer and a repressor (3). Comparisons between the wild-type and creBΔ strains showed that enzyme activity changes were mirrored at the transcript level in this organism as well. Importantly, this evidence indicates that the increased amount of expression, at least for the systems tested, has a transcriptional component and thus is likely due to a direct involvement of CreB in CCR rather than a consequence of the effects of creB on permeases (27). Although effects on permeases may alter intracellular inducer levels for some systems, e.g., at least in the case of ethanol induction of alcA in A. nidulans, no permease is required.
During our phenotypic analysis of the mutant strain, we found that glucose and, to a lesser degree, sucrose inhibit germination of A. oryzae RIB40 conidia. This inhibition of conidial germination was abolished in the creBΔ strain. Although we do not understand the molecular basis of this observation, this may provide a novel way of selecting new mutant strains: if spores are subjected to mutagenesis and then germinated in glucose-containing medium, the population of spores that regenerate is likely to be enriched for mutants with phenotypes overlapping those of the creBΔ strain. Furthermore, the germination of the creBΔ strain in the presence of glucose may be useful in industrial processes in which the preculture or primary fermentation is inoculated with conidia.
A. oryzae DAR3699 was already known to have many valuable properties for biotechnological use. Among 15 fungal strains tested, it had the highest protein content and shared the highest specific growth rate in starch processing wastewater, as well as having strong secreted amylase activity and a compact pellet morphology ideal for use in a bioreactor (14). In a pilot plant-scale air lift bioreactor, the strain efficiently converted carbon and other nutrients in starch processing wastewater into protein-rich fungal biomass (28). Yet little is known of the genetics of this strain. We have shown that A. oryzae DAR3699 has a mutation in the promoter of its creB gene; that it has many phenotypes consistent with loss of creB function, including strong growth on acetamide, weak growth on proline, quinate, or arabinose, resistance to molybdate, high protease secretion, and little or no inhibition of spore germination by glucose; and that deleting creB in this strain does not change the phenotype. A. oryzae arose through the ancient domestication of Aspergillus flavus and has undergone genetic changes during its centuries of use in Asian food and beverage production (29). Our data suggest that for A. oryzae DAR3699, one of these genetic changes is likely to have been partial or complete loss of creB function. This change may have been selected for the consequent increase in protease secretion, as this strain is used in soy fermentation, which is initially limited by low levels of free nitrogen.
In addition to being mutated in the model fungus A. nidulans (3), creB has now been mutated in three industrially useful fungi, diverse among the Ascomycetes: A. oryzae, P. decumbens (13), and T. reesei (12). In every case, loss of creB function has resulted in increased activities of multiple secreted hydrolases of industrial importance. Furthermore, we have shown that A. oryzae DAR3699, a strain useful in both solid-state food production and industrial wastewater treatment, is likely to have lost creB function. Taken together, these findings indicate that deletion of creB homologues may be broadly useful in a variety of fungi for producing a range of enzymes.
ACKNOWLEDGMENTS
A.J.H. was supported by an Australian Research Council linkage grant (LP0562153) awarded to B.J., C.P.S., and J.M.K. This work was cofunded by the South Australian Water Corporation (SA Water).
We thank Robin A. Lockington for his valuable input and Natasha T. Pyne for selection of the niaD mutant.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Ward OP, Qin WM, Dhanjoon J, Ye J, Singh A. 2006. Physiology and biotechnology of Aspergillus. Adv. Appl. Microbiol. 58:1–55 [PubMed] [Google Scholar]
- 2.Fleissner A, Dersch P. 2010. Expression and export: recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 87:1255–1270 [DOI] [PubMed] [Google Scholar]
- 3.Hynes MJ, Kelly JM. 1977. Pleiotropic mutants of Aspergillus nidulans altered in carbon metabolism. Mol. Gen. Genet. 150:193–204 [DOI] [PubMed] [Google Scholar]
- 4.Kelly JM, Hynes MJ. 1977. Increased and decreased sensitivity to carbon catabolite repression of enzymes of acetate metabolism in mutants of Aspergillus nidulans. Mol. Gen. Genet. 156:87–92 [DOI] [PubMed] [Google Scholar]
- 5.Abdallah BM, Simoes T, Fernandes A, Strauss J, Seiboth B, Sa-Correia I, Kubicek CP. 2000. Glucose does not activate the plasma-membrane-bound H+-ATPase but affects pmaA transcript abundance in Aspergillus nidulans. Arch. Microbiol. 174:340–345 [DOI] [PubMed] [Google Scholar]
- 6.Lockington RA, Kelly JM. 2001. Carbon catabolite repression in Aspergillus nidulans involves ubiquitination. Mol. Microbiol. 40:1311–1321 [DOI] [PubMed] [Google Scholar]
- 7.Lockington RA, Kelly JM. 2002. The WD40 repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol. Microbiol. 43:1173–1182 [DOI] [PubMed] [Google Scholar]
- 8.Dowzer CEA, Kelly JM. 1991. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 11:5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shroff RA, Lockington RA, Kelly JM. 1996. Analysis of mutations in the creA gene involved in carbon catabolite repression in Aspergillus nidulans. Can. J. Microbiol. 42:950–959 [DOI] [PubMed] [Google Scholar]
- 10.Shroff RA, O'Connor SM, Hynes MJ, Lockington RA, Kelly JM. 1997. Null alleles of creA, the regulator of carbon catabolite repression in Aspergillus nidulans. Fungal Genet. Biol. 22:28–38 [DOI] [PubMed] [Google Scholar]
- 11.Todd RB, Lockington RA, Kelly JM. 2000. The Aspergillus nidulans creC gene involved in carbon catabolite repression encodes a WD40 repeat protein. Mol. Gen. Genet. 263:561–570 [DOI] [PubMed] [Google Scholar]
- 12.Denton JA, Kelly JM. 2011. Disruption of Trichoderma reesei cre2, encoding an ubiquitin C-terminal hydrolase, results in increased cellulase activity. BMC Biotechnol. 11:103. 10.1186/1472-6750-11-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou G, Lü J, Li Z, Li J, Wang M, Qu Y, Xiao L, Qin S, Zhao H, Xia R, Fang X. 2012. Enhanced cellulase production of Penicillium decumbens by knocking out creB encoding a deubiquitination enzyme. Chin. J. Biotechnol. 28:959–972 [PubMed] [Google Scholar]
- 14.Jin B, van Leeuwen J, Yu Q, Patel B. 1999. Screening and selection of microfungi for microbial biomass protein production and water reclamation from starch processing wastewater. J. Chem. Technol. Biotechnol. 74:106–110 [Google Scholar]
- 15.Song MH, Nah J-Y, Han YS, Han DM, Chae K-S. 2001. Promotion of conidial head formation in Aspergillus oryzae by salt. Biotechnol. Lett. 23:689–691 [Google Scholar]
- 16.Schmoll M, Franchi L, Kubicek CP. 2005. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell 4:1998–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmoll M, Tian C, Sun J, Doris Tisch D, Glass NL. 2012. Unravelling the molecular basis for light modulated cellulase gene expression—the role of photoreceptors in Neurospora crassa. BMC Genomics 13:127. 10.1186/1471-2164-13-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda H, Sano M, Maruyama Y, Tanno T, Akao T, Totsuka Y, Endo M, Sakurada R, Machida M, Akita O, Hasegawa F, Abe K, Gomi K, Nakajima T, Iguchi Y. 2004. Transcriptional analysis of genes for energy catabolism and hydrolytic enzymes in the filamentous fungus Aspergillus oryzae using cDNA microarrays and expressed sequence tags. Appl. Microbiol. Biotechnol. 65:74–83 [DOI] [PubMed] [Google Scholar]
- 19.Kubodera T, Yamashita N, Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64:1416–1421 [DOI] [PubMed] [Google Scholar]
- 20.Ballance DJ, Turner G. 1985. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36:321–331 [DOI] [PubMed] [Google Scholar]
- 21.Hunter AJ, Jin B, Kelly JM. 2011. Independent duplications of alpha-amylase in different strains of Aspergillus oryzae. Fungal Genet. Biol. 48:438–444 [DOI] [PubMed] [Google Scholar]
- 22.Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T. 2009. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 85:141–154 [DOI] [PubMed] [Google Scholar]
- 23.Tada S, Iimura Y, Gomi K, Takahashi K, Hara Yoshizawa SK. 1989. Cloning and nucleotide sequence of the genomic taka-amylase A gene of Aspergillus oryzae. Agric. Biol. Chem. 53:593–599 [Google Scholar]
- 24.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K-I, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J-I, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 25.Rajkowitsch L, Vilela C, Berthelot K, Velasco Ramirez C, McCarthy JEG. 2004. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 335:71–85 [DOI] [PubMed] [Google Scholar]
- 26.Wada R, Maruyama J, Yamaguchi H, Yamamoto N, Wagu Y, Paoletti M, Archer DB, Dyer PS, Kitamota K. 2012. Presence and functionality of mating type genes in the supposedly asexual filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 78:2819–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamlangdee N. 2008. Identifying target proteins of the CreB deubiquitination enzyme in the fungus Aspergillus nidulans. Ph.D. thesis The University of Adelaide, Adelaide, Australia [Google Scholar]
- 28.Jin B, Yan XQ, Yu Q, van Leeuwen JH. 2002. A comprehensive pilot plant system for fungal biomass protein production and wastewater reclamation. Adv. Environ. Res. 6:179–189 [Google Scholar]
- 29.Gibbons JG, Salichos L, Slot JC, Rinker DC, McGary KL, King JG, Klich MA, Tabb DL, McDonald WH, Rokas A. 2012. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Biol. 22:1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]