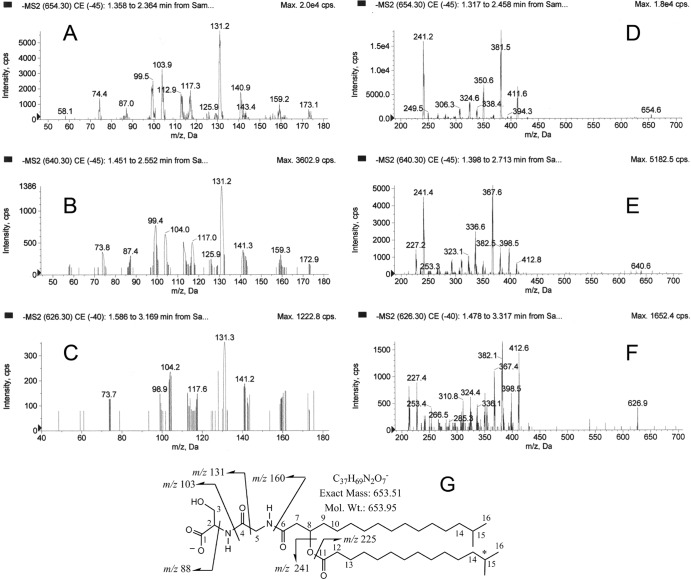
Fig 2.
MS/MS profiles of 654, 640, and 626 lipid species. A highly enriched preparation of lipid 654 was subjected to MS/MS analysis as described in Materials and Methods. The partial mass spectra are depicted for the m/z 654 lipid species (A and D), the m/z 640 lipid species (B and E), and the m/z 626 lipid species (C and F). The proposed structure of the most abundant species in the lipid 654 class is shown in graph G. The numbers adjacent to specific carbons refer to the NMR assignments shown in Table 1. The major low-mass product ions (<200 amu) detected with the three parent lipid species were essentially identical (A, B, and C), and these product ions represent cleavage products of the serine-glycine head group as depicted in the proposed chemical structure (G). Loss of the -CH2-OH from the m/z 103 fragment and addition of two protons yields an m/z 74 product ion. Loss of a proton from the m/z 88 fragment yields an m/z 87 product ion. Loss of the -CH2-OH from the m/z 131 fragment and loss of a proton yield an m/z 99 product ion. Loss of the -CH2-OH from the m/z 160 fragment and loss of a proton yield an m/z 129 product ion. Cleavage between carbons 7 and 8 and loss of the -CH2-OH group plus addition of two protons yield the m/z 173 ion product. The m/z 160 fragment forms an imine with loss of a proton to yield an m/z 159 product ion. The m/z 412 fragment results from loss of the C15:0 fatty acid (241 amu) from the 654 precursor ion. The m/z 381 fragment results from loss of C15:0 fatty acid and -CH2-OH from the 654 precursor ion. Fragmentation of the serine-glycine dipeptide together with loss of the C15:0 fatty acid results in the other ion fragments (m/z 250 to 420) depicted in graph D. Additional fatty acids described elsewhere in Results account for the 640 and 626 lipid species.