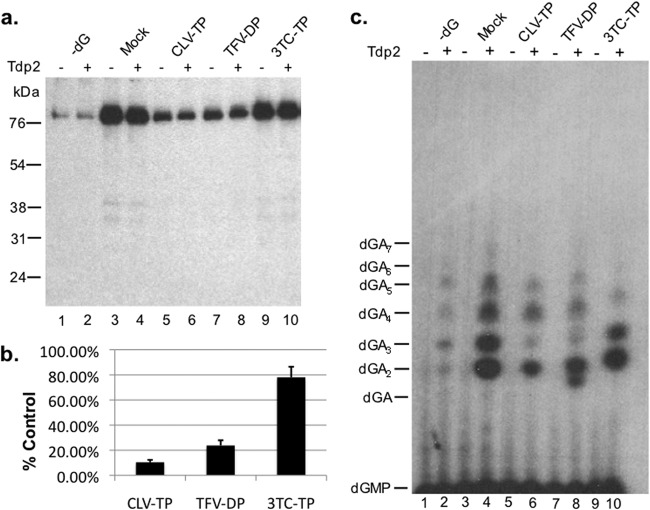
Fig 5.
Clevudine and tenofovir inhibited the DNA polymerization stage of HP protein priming in vitro. HP copurified with Hε in TMgNK buffer was primed with unlabeled dGTP (lanes 3 to 10). To monitor dGTP-independent (background) dATP incorporation, as described for Fig. 4, lane 1, no dGTP was added to the reactions shown in lanes 1 and 2 (−dG). Subsequently, samples were washed twice to remove unincorporated dGTP, and fresh TMgNK buffer was added along with dH2O (lanes 1 to 4) or 100 μM clevudine-TP (CLV-TP; lanes 5 and 6), tenofovir DF-DP (TFV-DP; lanes 7 and 8), or 3TC-TP (lanes 9 and 10). [α-32P]dATP was then added to all reactions to allow for polymerization in the presence or absence of potential drug inhibition. After priming, samples were washed extensively to remove unincorporated nucleotides and then were mock treated (odd-numbered lanes) or treated with Tdp2 (even-numbered lanes) to release DNA attached to HP. (a) The beads, which contained the primed HP, were processed for and resolved by SDS-PAGE and visualized by autoradiography. The positions of the protein molecular mass markers (in kDa) are indicated. (b) Priming signals (from samples not treated with Tdp2, i.e., the odd-numbered lanes in panel a) from three independent experiments were quantified by phosphorimaging, and after subtracting the background signal from lane 1 in panel a (−dG), they were expressed as percentages of the mock-treated control (lane 3). Error bars denote the standard errors of the means (SEM). (c) The supernatant, which contained the released DNA, was collected after Tdp2 digestion, resolved on an 8 M urea-20% polyacrylamide gel, and visualized by autoradiography. The positions of the dGAn (where n denotes the number of dA residues following the initial dG residue) oligonucleotide markers are indicated.