Fig 1.
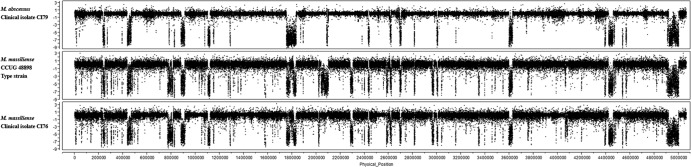
Whole-genome comparative genome hybridization (CGH) patterns of mycobacterial isolates relative to the M. abscessus type strain ATCC 19977. The normalized log2 signal ratios (y axis) for individual probes tiled across the genome (x axis) are shown for M. abscessus clinical isolate CI79 (top panel), the M. massiliense type strain CCUG 48898 (middle panel), and M. massiliense clinical isolate CI76 (bottom panel). Log ratios near 0 indicate genomic signal intensity similar to that of the M. abscessus reference genome, whereas contiguous regions of highly negative ratios indicate putative deletion segments (i.e., signal was absent due to lack of homologous DNA in the test strain).
