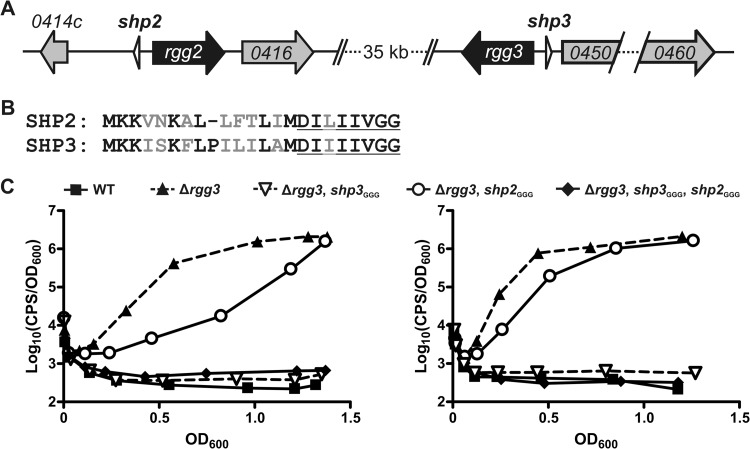
Fig 1.
Inactivation of shp genes differentially affects Rgg2/3 system autoinduction in the Δrgg3 background. (A) Organization of the rgg2 and rgg3 loci in the NZ131 genome. (B) Alignment of full-length prepeptides SHP2 and SHP3. Nonidentical amino acids are in gray font, and the C-terminal eight residues comprising the mature, active peptides are underlined. (C) Luciferase expression from Pshp2 reporter integrated into WT (BNL148), Δrgg3 (BNL149), Δrgg3 shp3GGG (BNL203), Δrgg3 shp2GGG (BNL176), and Δrgg3 shp3GGG shp2GGG (BNL199) isogenic mutant strains. Panels show results obtained with biological replicates using the same strains on separate occasions, and expression trends are representative of at least three independent experiments.