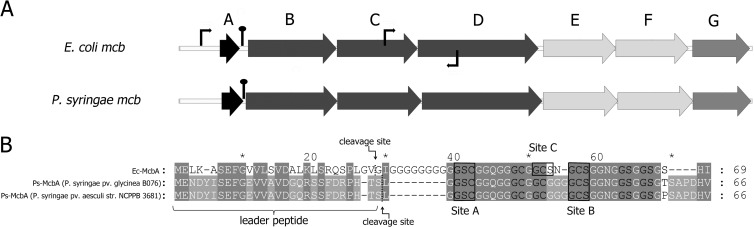
Fig 1.
Cluster of E. coli mcb gene homologs in P. syringae. (A) The E. coli mcbABCDEFG gene cluster is schematically presented at the top, with each gene of the cluster indicated by an arrow (not drawn to scale). The mcbBCD genes code for microcin B synthase, the mcbEF genes code for the microcin B efflux pump, and mcbG codes for a self-immunity protein. Thin arrows indicate known E. coli mcb promoters (31). The homologous operon found in some strains of P. syringae is shown below. The extent of similarity and identity between the E. coli and P. syringae homologs ranges from 63% and 46% (for the product of mcbC) to 45% and 25% (for the product of mcbG), respectively. Hairpin-like structures between mcbA and mcbB indicate putative transcription terminators located in noncoding regions (2). (B) Alignment of mcbA gene products from E. coli (Ec-McbA), P. syringae pv. glycinea (GenBank accession number NZ_AEGG01000085), and P. syringae pv. aesculi (accession number ACXS01000133) (single-letter amino acid codes). The degree of background shading indicates identity in all three (dark gray) or just two (lighter gray) of the aligned polypeptides. Hyphens indicate gaps. Ec-McbA di- and tripeptides converted into heterocycles (and homologous residues in P. syringae polypeptides) are indicated by black letters. Tripeptides converted to fused cycles (site A and site B) are boxed and labeled below the alignment. An Ec-McbA tripeptide converted into the auxiliary site C fused cycle is boxed and labeled above the alignment. The cleavage sites between the leader and the core parts of modified Ec-McbA and Ps-McbAs are highlighted above and below the alignment, correspondingly.