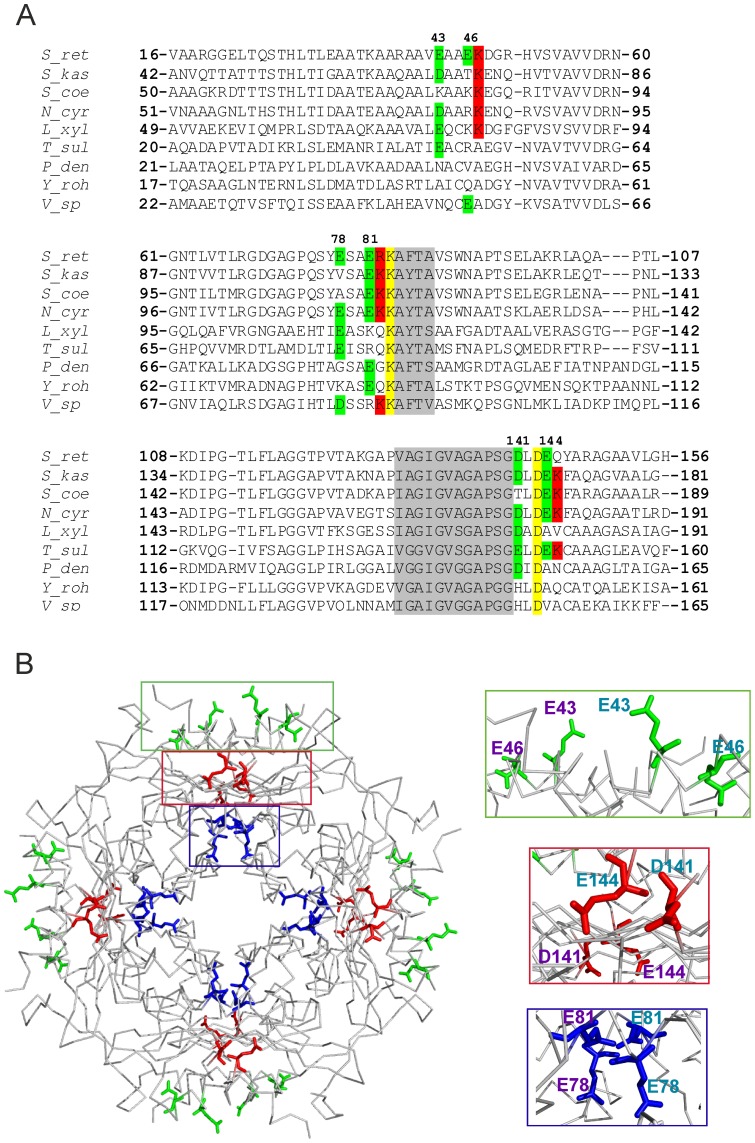
Figure 1. D/EXXE motifs within the HbpS sequence and their position on the octameric structure.
(A) Alignments of HbpS from S. reticuli (S_ret; GI:5834772; numbering according to PDB: 3FPV) and HbpS-like proteins from S. kasugaensis (S_kas; GI:157059904), S. coelicolor A3(2) (S_coe; GI: 8248773) Nocardia cyriacigeorgica GUH-2 (N_cyr; GI:379707916), Leifsonia. xyli subsp. xyli str. CTCB07 (L_xyl; GI: 50955378), Thioalkalivibrio sulfidophilus HL-EbGr7 (T_sul; GI: 220935915), Paracoccus denitrificans PD1222 (P_den; GI: 119383870), Yersinia rohdei ATCC 43380 (Y_roh; GI: 238750301) and Vibrio sp. MED222 (V_sp; GI: 86146209) are shown. Glutamates and aspartates from the studied D/EXXE motifs are marked with green background and their positions on HbpS are indicated. Conserved K83 and D143 are marked with yellow background. Neighbouring and conserved Lys and Arg are in red background. Conserved amino acid regions following E78XXE81 and preceding D141XXE144 are in grey background. (B) Positions of the exposed E43XXE46 (green box) and D141XXE144 (red box) as well as the internal E78XXE81 (blue box) on one 4-fold axis of the HbpS octamer are indicated (left). Enlargements of the marked boxes (right) are shown indicating the glutamates/aspartate from one subunit (E43, E46, E78, E81, D141 and E144 written in violet) and the adjacent subunit (Glu and Asp written in turquoise).