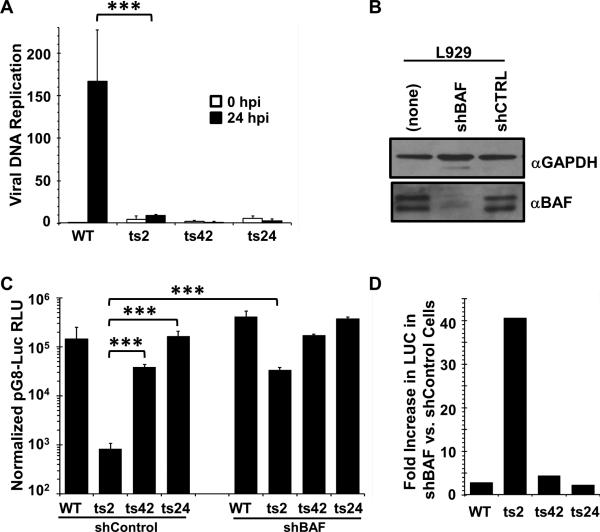
Figure 2. The defect in intermediate transcription is specific to ts2 and is rescued by depletion of BAF.
(A). DNA replication assays. Total DNA was isolated at 30 minutes and 24 hours after infection (MOI=3) with WT, ts2, ts42, or ts24 at 37°C, then quantified by qPCR. Data was obtained from two independent experiments, PCR amplified in duplicate. Data is shown as a fold difference compared to the WT sample at 0.5 hpi. Error bars represent standard deviation. (B). Immunoblot analysis of BAF expression in L929 cells stably depleted of BAF using specific shRNA. Lysates from equivalent numbers of cells were collected and analyzed using antibody against BAF. The total amounts of BAF in each lane were quantified using a Bio-Rad Chemidoc XRS instrument. GAPDH level was used as loading control. (C) L929 cells stably expressing shBAF or shControl were transfected with pG8-Luciferase for 7hr, then infected at 37°C with WT, ts2, ts42 or ts24 at MOI=3 in the presence of 50 μM AraC. Lysates were prepared at 16h after infection and assayed for luciferase activity. RLU shown is normalized to total protein measured by BCA assay. Data were obtained from three independent experiments performed in triplicate wells. Data from a representative experiment is shown. Error bars represent standard deviation. D) Data from the luciferase expression shown in (C) was replotted as a fold difference between the shControl and shBAF cell lines for each virus. (*** indicates a p-value <0.05)