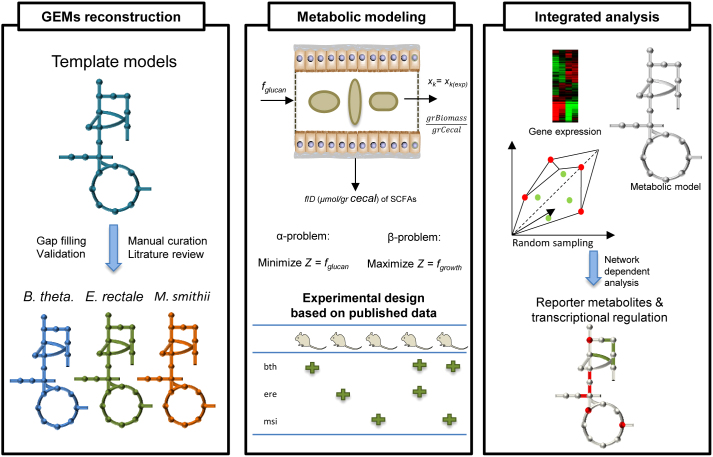
Figure 1. Genome-scale metabolic models (GEMs) for gut microbiota are reconstructed based on three template models and different databases.
This was followed by intensive manual curation of the GEMs for three key species in the human gut microbiome, based on literature and databases. The metabolic modeling step involved two approaches, the α- and β-problem which were defined depending on the availability of experimental data (the mono-colonization of germ free mice with B. thetaiotaomicron33, E. rectale33 and M. smithi47, the co-colonization of germ free mice with the B. thetaiotaomicron and E. rectale33, the B. thetaiotaomicron and M. smithii47). In the α-problem the abundance of the species were specified and the glucan and the SCFAs uptake and secretion profile was predicted. For the β-problem, the concentration of substrates in the feed was specified and the biomass and the SCFAs profile were predicted. Finally, the metabolic differences between each bacteria were identified through integrative analysis. Reporter metabolites, subnetworks and random sampling algorithms were used to reveal information about transcription regulations based on the transcriptome data.