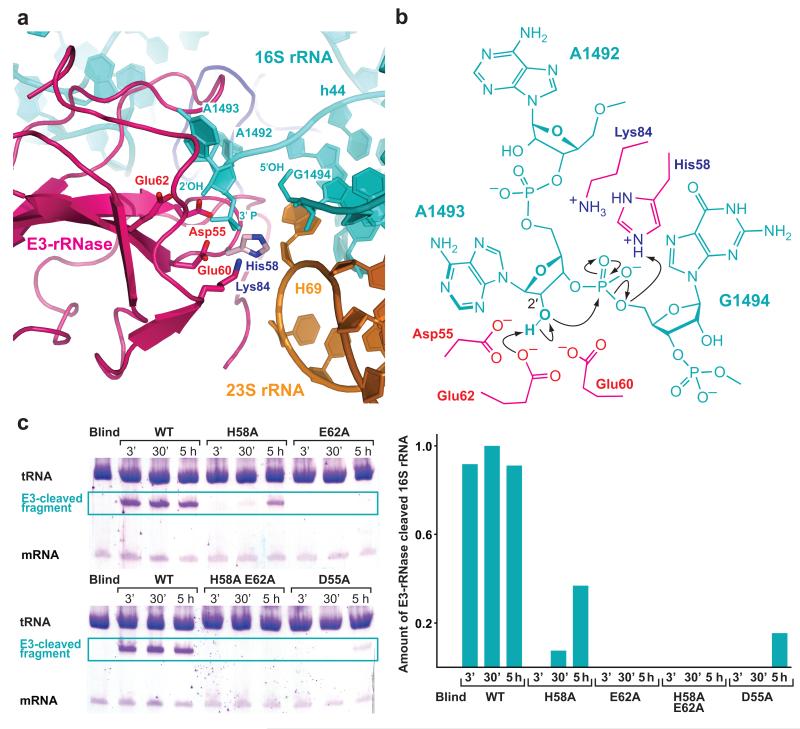
Figure 2. A mechanism for 16S rRNA cleavage by colicin E3-rRNase.
(a) The active site of E3-rRNase (pink cartoon) bound to the 70S ribosome with critical amino acids shown as labeled sticks. The structure represents the post-cleavage state, where the phosphodiester bond between A1493 and G1494 has been hydrolyzed, leading to a 3′-phosphate moiety at A1493 and a free 5′ OH at G1494. The E3-rRNase construct used in this study displayed a H58A mutation, residue His58, shown in light pink, has been modeled in this figure. (b) Schematic presentation of the proposed cleavage mechanism with residues from E3-rRNAse shown in pink. Glu62 deprotonates the 2′OH of A1493 activating it for a nucleophilic attack on the adjacent 3′-phosphate. The negatively charged trigonal bipyramidal transition state is likely to be stabilized by His58, which is also thought to finally protonate the leaving 5′-OH RNA fragment. (c) In vitro cleavage assay. E3-rRNase mutants (wt, H58A, E62A, H58A E62A, D55A) were incubated with a 10-fold excess of 70S Th. Thermophilus ribosomes, tRNAfMet and mRNA at 37°C. At the indicated time points the reactions were stopped by addition of phenol/chlorofom, the RNAs concentrated by ethanol precipitation and separated by 16% urea denaturating PAGE. The amounts of E3-cleaved 16S rRNA were determined by scanning the toluidine stained bands with a Typhoon imager and quantified with Image Quant TL (both GE Healthcare). While the wildtype E3-rRNase shows full cleavage activity already after 3 minutes, the mutants where either the proposed general base Glu62 or both the general acid and base (His58/Glu62) have been mutated to alanines decrease the E3-rRNase cleavage activity below the detection limit.