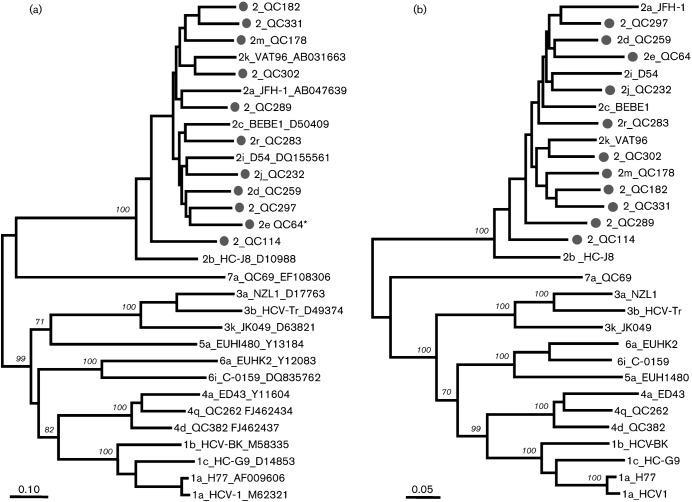
Fig. 1.
Maximum-likelihood phylogenies estimated from (a) full-length nucleotide sequences and (b) predicted amino acid sequences. Except for the 11 isolates from this study (•), reference sequences from subtypes 1a, 1b, 1c, 2a, 2b, 2c, 2i, 2k, 3a, 3b, 3k, 4a, 4d, 4q, 5a, 6a, 6i and 7a were also included. Each branch was led by an isolate named in the format: subtype_isolate_accession number. Isolate names that start with ‘2’ alone, without a letter, indicate that a subtype has not been assigned. Bootstrap supports are shown in italics. Bars, 0.10 nucleotide substitutions per site (a); 0.05 amino acid substitutions per site (b). *QC64 contains an extra p7 gene.