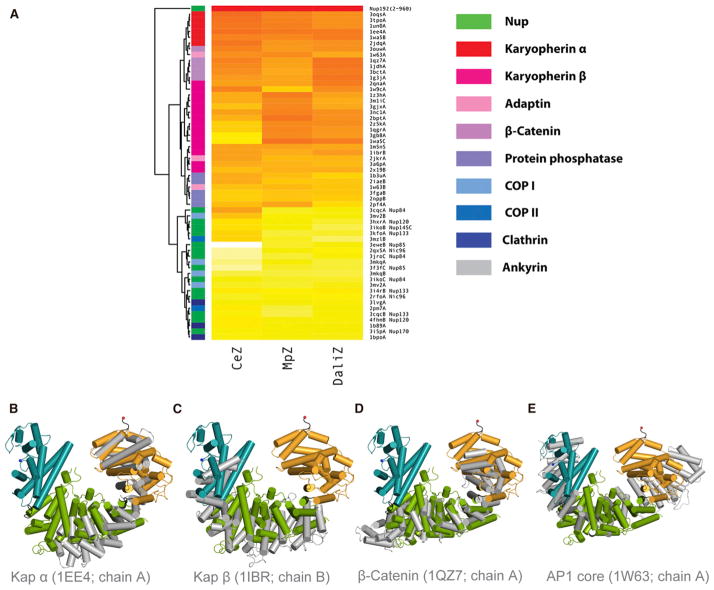
Figure 3. Structural Relatives of ScNup192(2–960).
(A) The heat map shown above illustrates the structural relationship of ScNup192(2–960) with selected α-helical proteins representing ten functional groups. Standardized scores for Dali, CE, and Multiprot alignments represented as a yellow to red gradient; red indicates stronger alignment scores (Table S2). The structure dendrogram is computed by hierarchical clustering using pairwise distances between alignment Z scores. The bar on the left shows the protein class for each structure, showing that karyopherins, β-catenin, and adaptin proteins are closest to Nup192. See also Figures S6B and S6C and Tables S4 and S5 for analyses on Nup85 and Nup170 structures.
(B–E) Structural superpositions of Nup192(2–960) with karyopherin 60 (PDB: 1EE4, chain A), karyopherin 95 (PDB: 1IBR, chain B), β-catenin (PDB: 1QZ7, chain A), and adaptor protein 1 (PDB: 1W63, chain A), respectively, are illustrated based on DALI alignments. These were the top structural alignment hits among the karyopherin alpha, karyopherin beta, β-catenin, and adaptin protein families. The D1, D2, and D3 domains of ScNup192(2–960) structure are shown in cyan, green, and gold, respectively, and the superposed molecules are in gray. See also Figure S6A.
See also Table S3.