Figure 1.
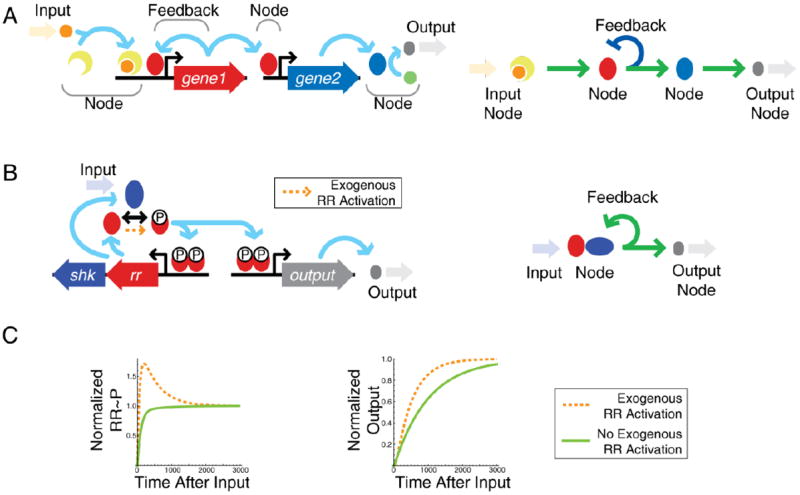
Information flow in signaling networks can strongly depend on non-transcriptional details with important physiological consequences. (A). Illustration of components in a transcriptional network. Input signals transfer information via nodes to create a physiological output. Right panel – transcriptional network diagram corresponding to detailed network on the left. C. A typical two-component system gene circuit is positively autoregulated by phosphorylated response regulator (RR). Right panel – simplified transcriptional network diagram corresponding to detailed network on the left. C. The system can exhibit feedback-induced overshoot (surge) kinetics if there is a small amount of regulator phosphorylation from an exogenous source (orange arrow and dashed line) in addition to sensor phosphorylation. In the absence of exogenous phosphorylation, induction is monotonic (solid green line). Overshoot of RR phosphorylation speeds induction of downstream genes (Normalized Output).
