Figure 5.
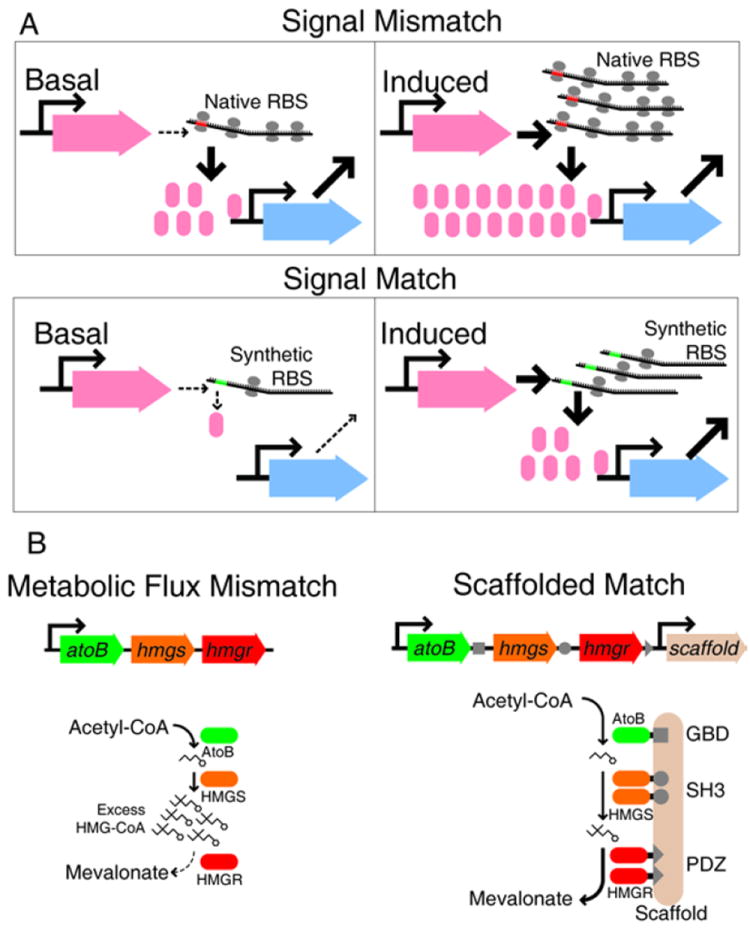
Engineering non-transcriptional processes for synthetic biology. A. ‘Leaky’ transcription can produce sufficient signal to flood downstream nodes. Signal matching can be achieved by engineering the ribosome binding site (RBS) to prevent low-level transcription from causing extraneous downstream signaling85. B. A similar mismatch occurs in a synthetic metabolic pathway for mevalonate production. A physical scaffold prevents accumulation of undesired intermediate HMG-CoA, reducing host cell toxicity and greatly increasing mevalonate product yield92, 93.
