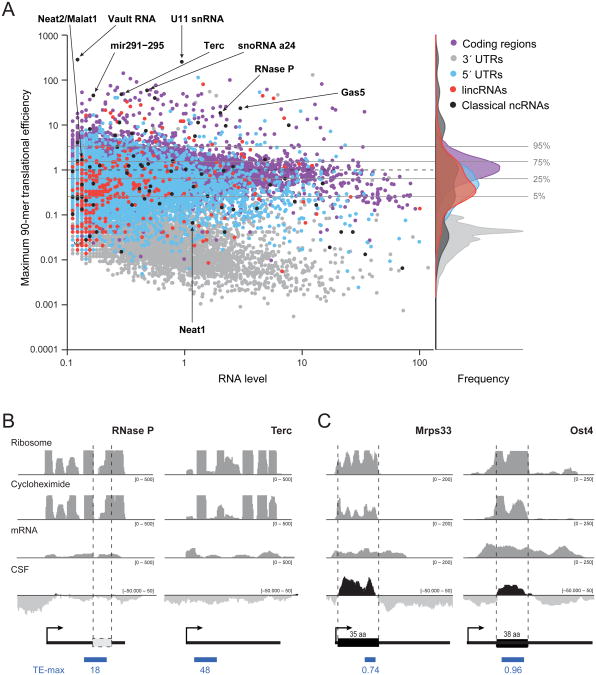
Figure 2. Translational efficiency of the maximum 90-mer fails to separate translated and non-translated RNAs.
(a) Scatter plot of RNA expression (log scale, x-axis) compared to the TE of the maximum 90-mer (log scale, y-axis) for coding regions (purple dots), 3′-UTRs (gray dots), 5′-UTRs (blue dots), classical ncRNAs (black dots), and lincRNAs (red dots). Horizontal lines correspond to the indicated percentiles of the TE-max score for protein-coding regions. The overlaid density distributions of the TE-max scores for each feature are shown. (b) Two examples of classical ncRNAs that have very high translational efficiency scores: RNase P and the telomerase RNA (Terc). The four rows (ribosome, cycloheximide, mRNA and CSF) are as described in legend of Figure 1. Beneath is an ideogram of the RNA, the location of a potential ORF (white box), and score of the maximum 90-mer (blue box). (c) Examples of two small coding genes encoding 35- and 38-amino acid peptides. See also Figure S2.