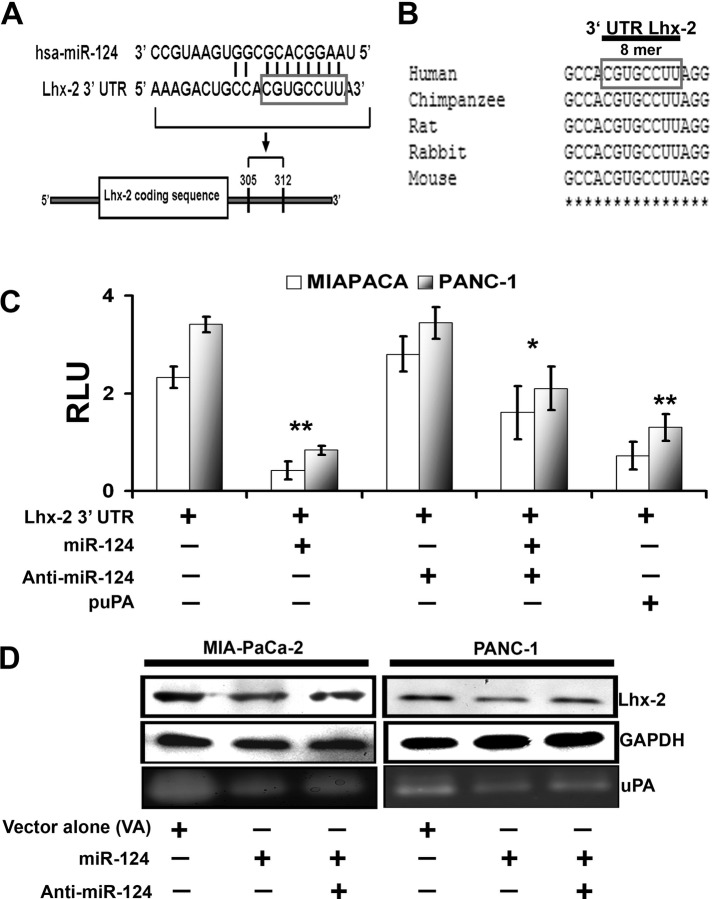
FIGURE 4:
Lhx2 is the predicted target for miR-124, which negatively regulates Lhx2. (A) Sequence alignment of miR-124 and predicted sequence pairing with a region of Lhx2 mRNA 3′-UTR. The nucleotides within the Lhx2 3′-UTR region that may interact with miR-124 are framed. (B) Alignment of nucleotide sequences of Lhx2 3′-UTR corresponding to the targets for miR-124 from several mammalian species. A high level of conservation suggests a functional role for these sequences. (C) Luciferase reporter assay. Interaction of miR-124 with Lhx2 3′-UTR luciferase reporter vector was transfected in the control and puPA-treated MIA PaCa-2 and PANC-1 cells alone and/or in combination with hsa-miR-124 and miR-124 inhibitor (anti–miR-124). Luciferase activity, which reflects extent of inhibition of Lhx2 3′-UTR reporter by miR-124, was quantified and normalized as described in Materials and Methods. The y-axis denotes relative luciferase units (RLU; mean ± SD; n = 3; *p < 0.05; **p < 0.01). (D) Western blot analysis of cell lysates (40 μg of total protein) obtained from control and hsa-miR-124– and/or anti–miR-124–treated MIA PaCa-2 and PANC-1 cells. Separated proteins were probed with anti-Lhx2 antibodies, followed by HRP-conjugated anti-rabbit secondary Abs and the bands were visualized with the chemiluminescent substrate. Anti-GAPDH Abs were used as a loading control. Fibrin zymography (bottom) was performed to determine uPA activity in the conditioned media of the cultured cells as described in Materials and Methods.