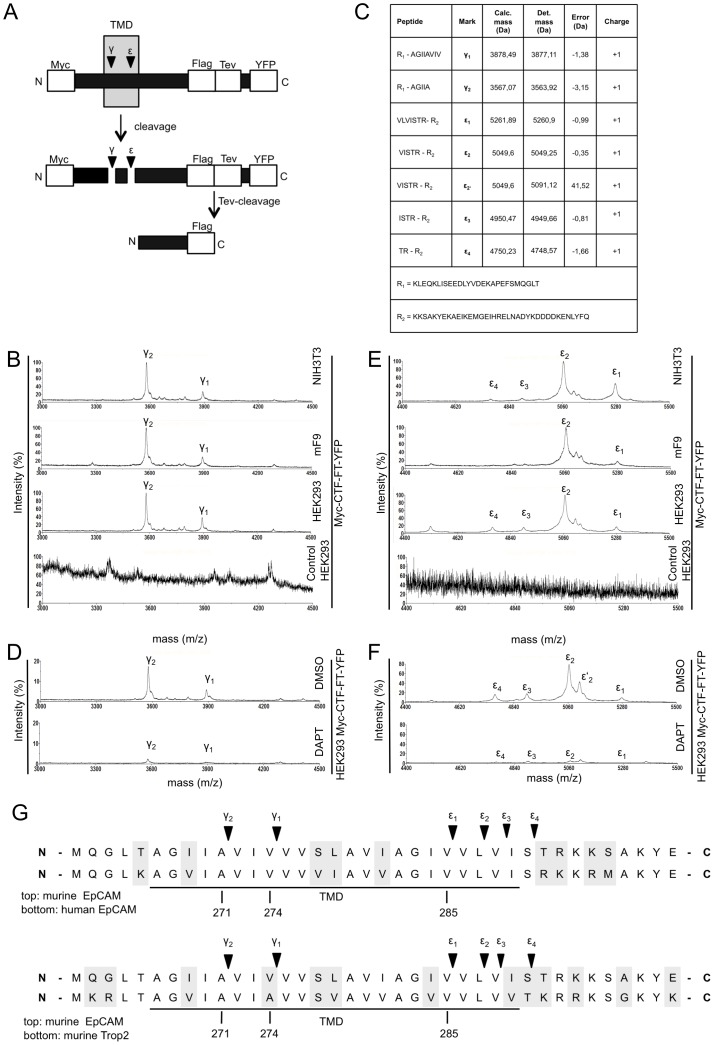
Figure 3. Determination of the γ-secretase cleavage amino acid sequence in mEpCAM.
(A) Schematic representation of Myc-CTF-YFP containing an N-terminal c-Myc-tag, a Flag-Tag and a TEV protease recognition site as well as YFP C-terminally of mEpICD. After cleavage by g-secretase, the Ab-like fragment can be isolated upon immunoprecipitation with c-Myc-specific antibodies. The YFP moiety is removed through digestion with TEV protease and the resulting small peptide isolated upon immunoprecipitation with Flag-specific antibodies. (B) Representative mass spectrometry spectrum of HEK293, NIH3T3, and mF9 cells stably expressing Myc-CTF-TF-YFP after immunoprecipitation of supernatants with c-Myc-specific antibodies. Two major peak species representing g-cleavages are indicated. (C) Tabular overview of γ-secretase cleavage sites within mEpCAM as determined upon mass spectrometric analysis and alignment to potential molecular weights. Calculated and determined masses are given in Dalton including error of each peptide. (D) Representative mass spectrometry spectrum of HEK293 cells stably expressing mEpCAM-TF after immunoprecipitation with c-Myc-specific antibodies and treatment with DMSO and the γ-secretase inhibitor DAPT. (E) Representative mass spectrometry spectrum of HEK293, mF9, and NIH3T3 stably expressing Myc-CTF-TF-YFP after TEV digestion and immunoprecipitation with Flag-specific antibodies. Four major peaks representing ε-cleavages are indicated. (F) Representative mass spectrometry spectrum of HEK293 cells stably expressing mEpCAM-TF after TEV digestion, immunoprecipitation with Flag-specific antibodies, and treatment with DMSO and the γ-secretase inhibitor DAPT. (G) Sequence alignment of murine and human EpCAM (top), and murine EpCAM and murine Trop-2 (bottom). γ-Secretase cleavages at g-position and e-position are indicated.