Abstract
In vitro, a new protocol of plant regeneration in rose was achieved via protocorm-like bodies (PLBs) induced from the root-like organs named rhizoids that developed from leaf explants. The development of rhizoids is a critical stage for efficient regeneration, which is triggered by exogenous auxin. However, the role of cytokinin in the control of organogenesis in rose is as yet uncharacterized. The aim of this study was to elucidate the molecular mechanism of cytokinin-modulated rhizoid formation in Rosa canina. Here, we found that cytokinin is a key regulator in the formation of rhizoids. Treatment with cytokinin reduced callus activity and significantly inhibited rhizoid formation in Rosa canina. We further isolated the full-length cDNA of a type-A response regulator gene of cytokinin signaling, RcRR1, from which the deduced amino acid sequence contained the conserved DDK motif. Gene expression analysis revealed that RcRR1 was differentially expressed during rhizoid formation and its expression level was rapidly up-regulated by cytokinin. In addition, the functionality of RcRR1 was tested in Arabidopsis. RcRR1 was found to be localized to the nucleus in GFP-RcRR1 transgenic plants and overexpression of RcRR1 resulted in increased primary root length and lateral root density. More importantly, RcRR1 overexpression transgenic plants also showed reduced sensitivity to cytokinin during root growth; auxin distribution and the expression of auxin efflux carriers PIN genes were altered in RcRR1 overexpression plants. Taken together, these results demonstrate that RcRR1 is a functional type-A response regulator which is involved in cytokinin-regulated rhizoid formation in Rosa canina.
Introduction
Rose (Rosa spp.) is one of the most important and widely grown ornamental plants in the world [1]. Previous studies have shown that rose can be efficiently regenerated through organogenesis from various explants, such as leaves, roots, internodes, petioles and immature seeds, which is a prerequisite for plant reproduction and genetic engineering [2], [3], [4]. Increasing evidence has demonstrated that a precise balance of exogenous hormones is required for the organogenesis. We previously established that a certain concentration of auxin is essential for the formation of the root-like organs named rhizoids from leaf explants in Rosa canina [5], [6]. However, the role of cytokinin in this process has not been characterized.
Cytokinins are a class of plant hormones that were first identified by their role, in concert with auxin, in the control of cell division in cultured plant cells [7], [8], [9]. While besides their ability to stimulate cell division, cytokinin regulates diverse aspects of plant growth and development, including embryo and seed development [10], [11], shoot initiation [12], leaf senescence [13], vascular differentiation [14] and response to biotic and abiotic stresses [15], [16]. Because of the biological and agricultural importance of cytokinin [17], [18], it is important to understand the basic mechanism by which they regulate such diverse processes in plant cells. In the past decade, substantial progress has been made in characterizing the cytokinin signal transduction pathway in Arabidopsis, including the identification of the genes encoding the proteins controlling the key steps in cytokinin biosynthesis and signaling [19], [20], [21].
Cytokinin signal transduction is based on the two-component phosphorelay signal transduction pathway which mainly includes Histidine Kinases (HKs), Histidine Phosphotransfer Proteins (HPs) and Response Regulators (RRs) [22], [23], [24]. In Arabidopsis, response regulators (ARRs) fall into four main groups on the basis of their sequence similarities, domain structure and transcriptional response to cytokinin, including type-A ARRs, type-B ARRs, type-C ARRs and Arabidopsis pseudo-response regulators (APRRs) [25], [26]. Type-A ARRs have a short C-terminal sequence, but lack the DNA-binding domain named GARP [27], [28]. In contrast to type-A ARRs, type-B ARRs contain a receiver domain and a long C-terminal extension carrying a DNA-binding domain that is responsible for the regulation of transcription of cytokinin-related genes [29]. Type-C ARRs share less sequence similarity to type-A and type-B ARRs; they do not contain DNA-binding domains and are not transcriptionally regulated by cytokinin [30]. The APRRs lack the conserved residues for phosphorylation and are involved in modulating circadian rhythms [31]. However, the response regulators have not been characterized in ornamental plants.
In this study, we found that cytokinin treatment significantly reduced callus activity and rhizoid formation in Rosa canina. Furthermore, we identified a type-A response regulator homologous gene, RcRR1, and found that its expression was rapidly induced by cytokinin. Importantly, RcRR1 overexpression Arabidopsis plants showed reduced sensitivity to cytokinin and altered auxin distribution and PIN genes expression. These findings provide new insights into the mechanism of cytokinin signaling in regulation of organogenesis in Rosa canina.
Results
Cytokinin Modulates Rhizoid Organogenesis
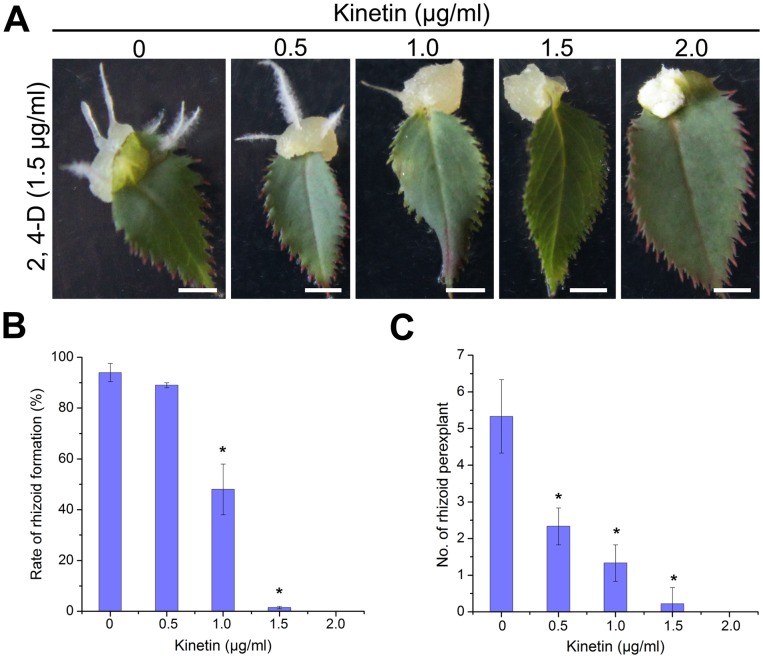
We have established a new plant regeneration system using protocorm-like bodies (PLBs) that can be induced from the tips of rhizoids and adapted this system to further study the effects of cytokinin on rhizoid formation in Rosa canina. As shown in Figure S1, leaf explants of Rosa canina were cultivated in Murashige and Skoog (MS) medium containing cytokinin and auxin in various concentrations. The rhizoids could be induced with appropriate auxin concentration even without exogenous cytokinin, while at high levels of auxin, the ability of rhizoid formation decreased (Figure S1). However, we observed a decreased ability of leaf explants to form rhizoids even at low cytokinin levels. With the increasing of cytokinin concentration in the medium, the callus became yellow and gradually disorganized with rare distinguishable root-like structures (Figure 1A and 1B).
Figure 1. Cytokinin modulates auxin-induced rhizoid organogenesis.
(A) Induction of rhizoids by 1.5 µg/ml 2, 4-D at various concentrations of kinetin. Scale bar, 2 mm. (B) Rate of rhizoid formation. (C) Number of rhizoids per explant. Asterisks in (B) and (C) indicate statistically significant differences (Student’s t-test; P<0.01) between the control and cytokinin-treated leaf explants; error bars show SDs.
Statistical analysis further showed that the number of root-like structures formed per explant decreased 50% at 0.5 µg/ml cytokinin, and the formation of root-like structures was totally inhibited at 2 µg/ml cytokinin (Figure 1C). These results suggest that auxin alone is able to trigger the formation of new organs and that cytokinin plays a regulatory role in organogenesis and ultimately plant regeneration.
Cloning and Characterization of RcRR1
To address the regulatory role of cytokinin in rhizoid formation during Rosa canina regeneration, we cloned a type-A ARR homologous gene. A cDNA fragment was amplified using degenerate primers and then the full-length cDNA (accession number KC952001) named RcRR1 was isolated by rapid amplification of cDNA ends (RACE). The full transcript of RcRR1 is 939 bp in length and contains a 702 bp open reading frame, a 116 bp 5′-untranslated region (UTR) and a 121 bp 3′-UTR.
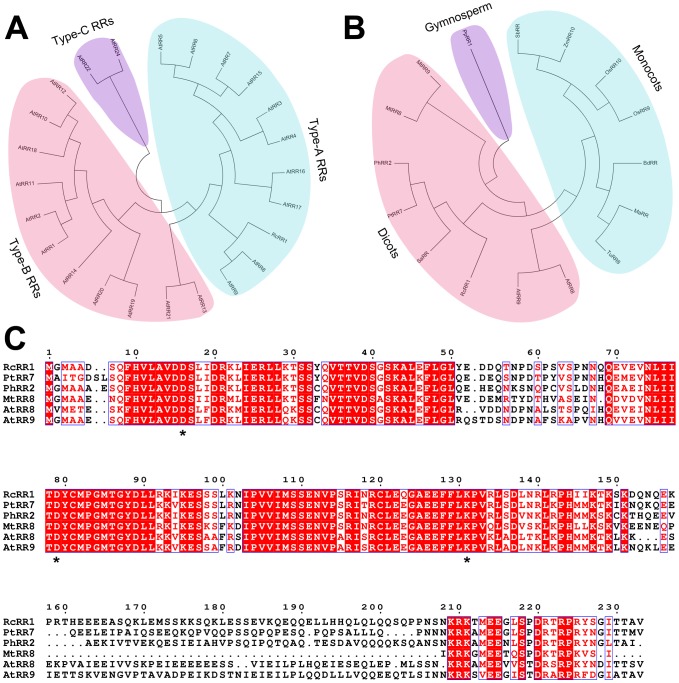
To characterize the function of RcRR1 in Rosa canina, we performed phylogenetic analysis using MEGA4. Our analysis showed that RcRR1 is more closely related to type-A response regulator genes than to other types in Arabidopsis (Figure 2A). In addition, RcRR1 was closely related to type-A ARRs from dicots (Figure 2B). Sequence alignment analysis further showed that RcRR1 contains all the features of a type-A response regulator (including the conserved DDK residues and a short C-terminal domain) and shares high similarity with PtRR7 from Populus trichocarpa (69% identity), PhRR2 from Petunia x hybrid (68% identity), ARR8 from Arabidopsis (59% identity), ARR9 from Arabidopsis (60% identity) and MtRR8 from Medicago truncatula (58% identity) (Figure 2C).
Figure 2. Sequence analysis of the full-length cDNA encoding a Rosa canina type-A response regulator, RcRR1.
(A) Phylogenetic analysis of putative RcRR1 from Rosa canina and response regulators in Arabidopsis. The aligned sequences were used to construct a phylogenetic tree by MEGA4. (B) Phylogenetic analysis of response regulators from a range of plant species. (C) Sequence alignment of RcRR1 homologs from different species. The alignment was generated using ClustalW and Espript (http://espript.ibcp.fr/ESPript/ESPript/).
RcRR1 Expression is Regulated by Cytokinin
We determined the tissue-specific expression pattern of RcRR1 by RT-PCR analysis. As shown in Figure S2, the highest level of RcRR1 expression was seen in roots, suggesting that this gene has a functional importance in roots (Figure S2). Next, we analyzed the expression of RcRR1 over the course of rhizoid development. Our results showed that there was high expression of RcRR1 at the early stage of callus formation, and the peak expression level was reached during rhizoid formation, after which the expression of RcRR1 decreased gradually (Figure S3). These results indicate that RcRR1 expression has a strong correlation with rhizoid development process.
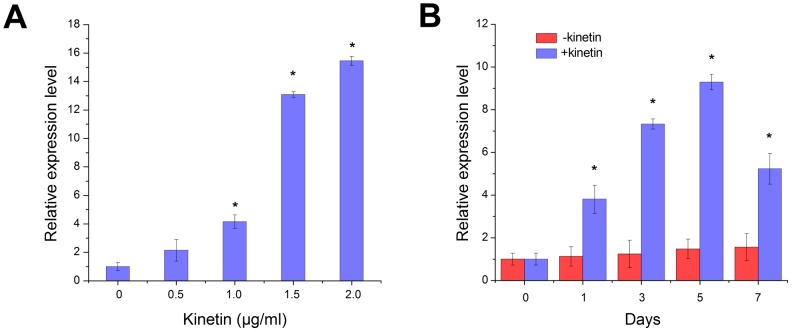
We investigated the effects of exogenous cytokinin treatment on RcRR1 expression after 1 week of culture on rhizoid formation medium. As the kinetin concentration increased, the level of RcRR1 expression was significantly up-regulated as compared with the control group (Figure 3A). We then examined RcRR1 expression in leaf explants on a short time-scale with 0.5 µg/ml kinetin treatment in order to understand how quickly was RcRR1 response to cytokinin. Our results showed that the expression of RcRR1 was rapidly up-regulated upon the cytokinin treatment, whereas the expression of RcRR1 was almost unchanged in the untreated explants (Figure 3B).
Figure 3. Quantitative real-time PCR shows regulation of RcRR1 expression by cytokinin.
(A) RcRR1 expression in explants incubated on medium containing 0, 0.5, 1.0, 1.5 and 2.0 µg/ml kinetin for 7 days. (B) Expression of RcRR1 in response to 0.5 µg/ml kinetin at various incubation times. Asterisks in (A) and (B) indicate statistically significant differences (Student’s t-test; P<0.01) between the control and cytokinin-treated leaf explants; error bars show SDs.
We additionally tested the induction of RcRR1 in callus by a variety of stress treatments. Besides cytokinin, the expression level of RcRR1 was not affected by any stresses used in this experiment, suggesting that RcRR1 is specific response to cytokinin (Figure S4). To examine whether cytokinin affects the expression of auxin-related gene, we also homology cloned a Rosa canina expressed sequence tag (EST) of the auxin efflux carrier gene from rhizoids and investigated the effect of cytokinin on its expression. RT-PCR showed that the transcription of the putative auxin efflux carrier gene dramatically decreased with increasing cytokinin concentrations (Figure S5).
RcRR1 Is Localized in the Nucleus
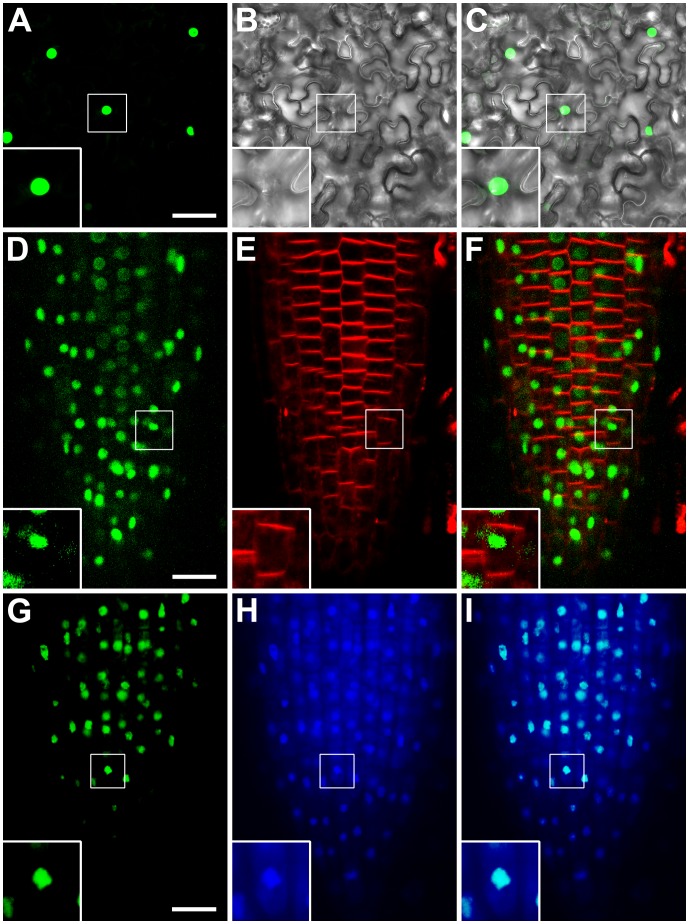
To further characterize the function of RcRR1, we next investigated the subcellular localization of RcRR1. We used different strategies, including transient expression analysis in Nicotiana benthamiana and stable Arabidopsis lines transformed with 35S: GFP-RcRR1. We found that in leaf cells of transiently transformed tobacco, the green fluorescent protein-tagged RcRR1 (GFP-RcRR1) was localized in the nucleus (Figure 4A–C). In transgenic Arabidopsis, GFP-RcRR1 also showed a high fluorescent signal as speckles in the nucleus (Figure 4D–F); these results were confirmed through colocalization analysis using the nucleus-specific dye, 4', 6-diamidino-2-phenylindole (DAPI) (Figure 4G–I). All these results suggest that GFP-RcRR1 is a nuclear protein and may act as a transcriptional regulator.
Figure 4. Subcellular localization of RcRR1.
(A–C) Localization of GFP-RcRR1 in leaf cells of tobacco. (D–I) Localization of GFP-RcRR1 in Arabidopsis root cells incubated with FM4-64 (D–F) and DAPI (G–I). Imaging of the 35S::GFP-RcRR1 fusion protein was conducted on a laser scanning confocal microscope. Insets show higher magnification views. Scale bar, 50 µm in (A–C) and 20 µm in (D–I).
Characterization of RcRR1 Overexpression Transgenic Arabidopsis
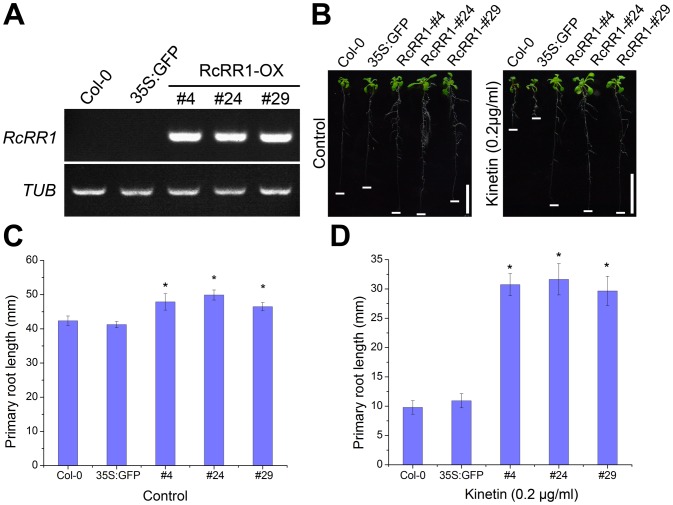
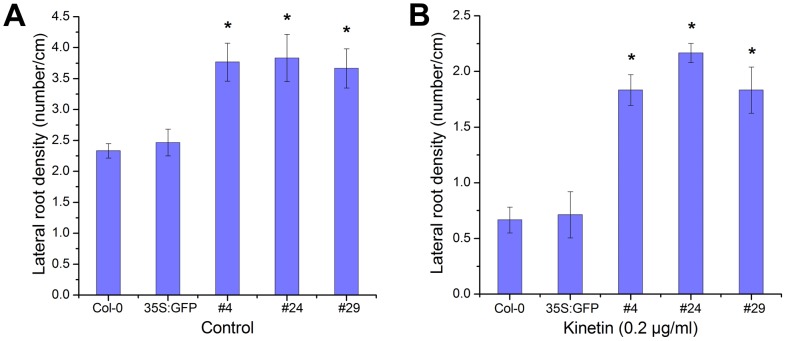
We functionally characterized the role of RcRR1 in root formation by overexpression of RcRR1 in Arabidopsis. The cDNA fragment containing coding sequences of RcRR1 was fused in-frame to a GFP sequence, and the GFP-RcRR1 fusion gene was placed under the control of a cauliflower mosaic virus (CaMV) 35S promoter. All of these transgenic lines showed high expression levels of GFP-RcRR1 as revealed by RT-PCR (Figure 5A). The RcRR1-OX transgenic plants had a longer primary root than that of the wild type (Figure 5B). Further statistical analysis revealed that the transgenic line #24 had the longest primary root (16% longer than that of wild-type plants) (Figure 5C). As shown in Figure 6A, RcRR1 overexpression also resulted in significantly increased lateral root numbers as compared with wild-type plants. The #24 transgenic plants showed the strongest phenotype with 39% more lateral roots than wild-type plants (Figure 6A).
Figure 5. Overexpression of the RcRR1 gene in Arabidopsis.
(A) Analysis of RcRR1 transcripts by RT-PCR in the wild-type and 35S::GFP-RcRR1 transgenic plants, respectively. TUBULIN (TUB) was used as an internal control. (B) Phenotypes of 10-day-old seedlings of wild-type and transgenic plants treated with or without 0.2 µg/ml kinetin. Scale bar, 1 cm. (C) Statistical analysis of primary root length of wild-type and transgenic plants. (D) Statistical analysis of primary root length of wild-type and transgenic plants treated with 0.2 µg/ml kinetin. Asterisks in (C) and (D) indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
Figure 6. Overexpression of RcRR1 affects the density of lateral root in transgenic Arabidopsis.
(A) Statistical analysis of the number of lateral roots (LR) per centimeter of the primary root from the wild-type and transgenic plants. (B) Statistical analysis of the number of LR in the wild-type and transgenic plants treated with 0.2 µg/ml kinetin. Asterisks in (A) and (B) indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
In addition, we also investigated the sensitivity of RcRR1-OX plants to cytokinin. We found that the primary root length, lateral root density and primary root elongation rate of transgenic plants showed varying degrees of reduced sensitivity to cytokinin in comparison with the wild type (Figure 5B; Figure S6 and S7). Treatment with 0.2 µg/ml kinetin caused more than 79% inhibition of primary root length (Figure 5C and 5D) and 71% decrease of lateral root numbers (Figure 6A and 6B) in wild-type plants; however, this dose had a significantly lower effect on the primary root length and lateral root density of RcRR1-OX lines. Of these three lines, the #24 transgenic plants showed the strongest phenotype, with primary root length and lateral root density reduced by 40% and 45%, respectively (Figure 5C and 5D; Figure 6A and 6B).
Auxin Signaling is Altered in the RcRR1-OX Arabidopsis Root
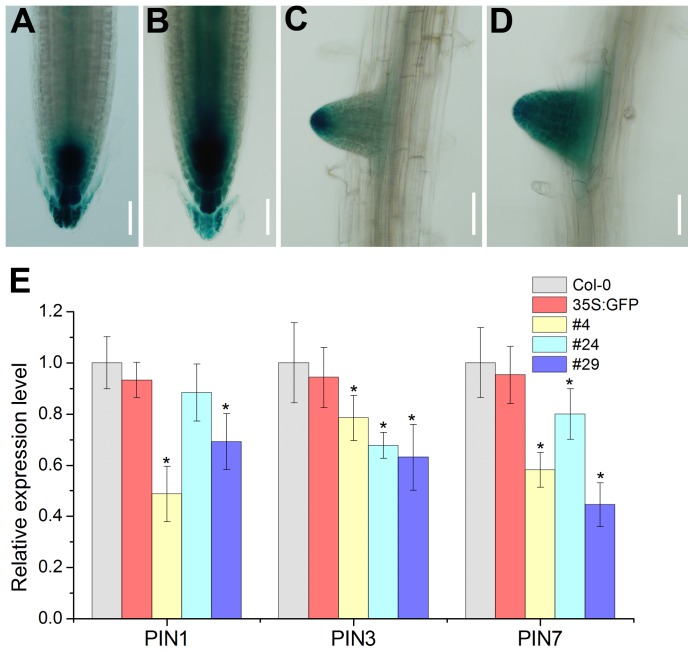
DR5::GUS is a useful reporter gene, which is widely used for visualizing the distribution of auxin [32], [33]. To investigate whether RcRR1-OX affects the distribution of auxin in Arabidopsis root, the DR5::GUS reporter was transformed into wild-type plants and hybridized with T2 seedlings of RcRR1-OX Arabidopsis. In the wild type, DR5::GUS staining was observed in the root cap and quiescent center (Figure 7A). In contrast, DR5::GUS showed a wider distribution in RcRR1-OX plants which extended to the pericycle and endodermis (Figure 7B). The broadening distribution of auxin was also observed in the lateral roots of RcRR1-OX plants (Figure 7C and 7D). We further studied the effect of RcRR1-OX on the expression of the PIN1, PIN3 and PIN7 genes. As shown in Figure 7E, the expression of these three genes was significantly down-regulated in the RcRR1-OX Arabidopsis in comparison with the wild type (Figure 7E). Our results suggest that RcRR1 might be correlated with the auxin signaling to control root organogenesis.
Figure 7. Auxin signaling is altered in the RcRR1-OX Arabidopsis.
(A, B) DR5::GUS expression in primary root tips of 6-day-old seedlings of wild-type (A) and transgenic (B) plants. Scale bar, 50 µm. (C, D) DR5::GUS expression in lateral roots of 6-day-old seedlings of wild-type (C) and transgenic (D) plants. Scale bar, 50 µm. (E) Expression of auxin efflux carrier genes in the wild-type and transgenic plants. Ubiquitin (UBQ) was used as an internal control.
Discussion
Rapid and efficient regeneration of rose in vitro is very important for rose reproduction. We have reported a new way of plant regeneration via protocorm-like bodies (PLBs), which were induced from rhizoids developed from leaf explants of Rosa canina [6]. It is well known that in vitro plant regeneration via embryogenesis or organogenesis is dependent on the hormone composition of the culture medium [34]. Although it has been proved that auxin is essential for rhizoid initiation, the role of cytokinin in this process remained unclear. It has been reported that the phytohormone cytokinin modulates multiple plant developmental processes, including cell division and differentiation [35], [36], apical dominance [37], [38] and light responses [39]. In this study, we found that cytokinin treatment alone was unable to trigger any organogenic response but reduced callus activity. Moreover, cytokinin significantly inhibited the auxin-induced rhizoid formation. Yet, we can’t explain the precise mechanism of the link between callus activity and rhizoid output. The most probable explanation is that cytokinin treatment changed hormone balance in callus cells, reduced callus activity and ultimately inhibited rhizoid formation. These findings suggest that cytokinin is a key regulator of rhizoid development.
Type-A ARRs have been shown to participate in various cytokinin-related plant growth and development processes [40], [41]. In Arabidopsis, ARR4 overexpression confers hypersensitivity to red light, indicating a role in light-regulated development [42]. Overexpression of a rice type-A ARR gene also results in hyposensitivity to cytokinin in callus formation [43]. In the present study, we isolated a putative response regulator gene named RcRR1 and found that the deduced amino acid sequence of RcRR1 showed high similarity to the type-A ARRs in plants and contained the conserved DDK domain indicating that RcRR1 belonged to the type-A ARRs subfamily. Phylogenetic analysis further showed that the response regulator genes were distributed in a wide range of plants including dicots, monocots and gymnosperms, indicating their important function in various species.
In Arabidopsis, transcription of most of the type-A ARRs has been detected in all adult organs, and the highest levels are found in the root [44]. In this study, we found that RcRR1 was also mainly expressed in the root, followed by the stem and leaves. This high expression of RcRR1 in the root indicates that it may have a function during root-organ formation; moreover, there was a high expression level of RcRR1 at an early stage of callus formation, which was closely correlated with the rhizoid formation process, suggesting that RcRR1 is involved in rhizoid organogenesis.
The expression of some type-A ARRs genes is regulated by various environmental stresses, nitrogen levels and phosphate starvation [45], [46], [47]. Our study showed that cytokinin substantially increased the transcript level of RcRR1. The up-regulating effect of cytokinin on RcRR1 expression was observed after short-term treatment, suggesting that RcRR1 expression can be rapidly regulated by cytokinin, which is consistent with the results in Arabidopsis, M. truncatula, and P. trichocarpa. Additionally, the development of primary and lateral roots of RcRR1 overexpression transgenic Arabidopsis showed reduced sensitivity to cytokinin, further demonstrating that RcRR1 is a functional negative response regulator of cytokinin signaling.
The interaction between auxin and cytokinin in plant organogenesis has been well-established [48], [49], [50]. However, the molecular mechanism of their interaction in controlling plant organogenesis remains uncharacterized. Pernisová et al. found that cytokinin modulates auxin-induced root-like organs in Arabidopsis via regulation of the auxin efflux [51]. PIN1, PIN3 and PIN7 have been identified as the principal root-specific auxin efflux carriers controlling the auxin distribution during root development [33]. We found that the expression pattern of DR5 was dramatically changed in the primary root tip and lateral roots of the RcRR1-OX plants. Furthermore, the expression of PIN1, PIN3 and PIN7 genes was down-regulated in RcRR1-OX Arabidopsis as compared with the wild type. In addition, the expression of the putative auxin efflux carrier gene in Rosa canina was decreased under cytokinin treatment. Taken together, these results indicate that cytokinin might be associated with the auxin signaling in controlling rhizoid organogenesis via RcRR1 in Rosa canina.
Materials and Methods
Plant Materials and Growth Conditions
Rosa canina was used in this study. Stem segments from 1-year-old shoots were disinfected according to Tian et al. (2008) [6]. Single node segments were cultured on MS media supplemented with 1.0 mg/l 6-benzyladenine (6-BA), 0.004 mg/l a-naphthalene acetic acid (NAA) and 0.1 mg/l gibberellic acid (GA3) at 25°C with a 16 hr light/8 hr dark regimen and a light intensity of 50 mEm−2/s. Leaves of 6-week-old plantlets were collected and placed with their adaxial sides down onto rhizoid formation medium as described previously.
Cytokinin Treatment
To investigate the role of cytokinin in the formation of rhizoids, the leaf explants were cultured on rhizoid formation medium containing 0, 0.5, 1.0, 1.5 and 2.0 µg/ml kinetin. Rhizoid formation was analyzed after 3 weeks of culture.
Isolation of the Full-length Coding Sequence for RcRR1
RNA was extracted from the callus with rhizoids of Rosa canina using RN09-EASY spin Kit (Biomed, China) and digested with RNase-free DNaseI. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, USA). Primers were designed for RcRR1 cloning based on the sequences regions of type-A ARRs genes that were conserved among Arabidopsis, P. trichocarpa and M. truncatula. After obtaining a partial fragment using forward primer P_for and reverse primer P_rev, the 3′ and 5′ sequence of RcRR1 were amplified by the RACE method. The RcRR1 full-length cDNA was isolated by combining the above three fragments. The products amplified using PrimeStarHS DNA Polymerase (Takara, Japan) were cloned into the pEASY-Blunt Simple Cloning Vector (Trans, China) and verified by sequencing (Sangon Biotech, Shanghai, China). Primers sequences used for the constructs are listed in Table S1. Sequence alignment and phylogenic analysis were performed using the ClustalW, ESPript programs (http://www.genome.jp/tools/clustalw/) and MEGA4 program. Accession numbers used for the phylogenic analysis are listed in Table S2.
RNA Preparation and Real-Time Quantitative PCR Analysis
For RcRR1 transcripts analysis, total RNAs were extracted from Rosa canina leaf explants treated with various concentrations of kinetin for various lengths of time using the RN09-EASY spin Kit (Biomed, China). The DNaseI-treated RNA (2 µg) of each tissue was reverse transcribed using Superscript III reverse transcriptase (Invitrogen, USA). Real-time Quantitative PCR was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems, USA) using SYBR® Premix Ex Taq™ II (TaKaRa, Japan). The reaction procedure was as follows: denature at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C, 34 s at 58°C. The Rosa canina 18S rRNA gene was used as an internal control for normalization, and the data were analyzed by 7500 Software version 2.0.4. The relative expression of the detected genes was calculated using the relative 2−ΔΔCT method [52]. Primers used for qRT-PCR are listed in Table S1.
Vector Construction and Plant Transformation
The open reading frame (ORF) of RcRR1 was amplified with primers PstI_for and BamHI_rev. After digestion with BamHI and PstI, the fragment was inserted into pCambia 2300-GFP to obtain the p2300-GFP-RcRR1 expression vector for tobacco and Arabidopsis transformation. Tobacco leaves were transiently transformed via A. tumefaciens leaf infiltration according to Batoko et al. (2000) [53]. Arabidopsis were infiltrated with Agrobacterium using the floral-dip method [54].
The DR5::GUS expression vector was constructed as follows: the DR5 promoter was amplified from the DR5::GFP plasmid and subcloned into pBI121-GUS to produce the DR5::GUS expression vector. Primer sequences used for the constructs are listed in Table S1.
Cytokinin Response Analysis
For the assay of primary root length and lateral root density, Arabidopsis seedlings were grown vertically on 1/2 MS medium supplemented with appropriate concentrations of kinetin. Root length was measured 10 days after germination using Image J software (http://rsb.info.nih.gov/ij/). The lateral root number was counted and converted into the number of lateral roots per centimeter of the primary root.
Microscopy
For analysis of the subcellular localization of RcRR1, transiently transformed tobacco leaves were cut out, mounted onto glass slides, and viewed under confocal laser scanning microscope (CLSM). Stable transformed GFP-RcRR1 Arabidopsis seedlings were incubated with FM4-64 (red) and DAPI (blue) for 3 min and 15 min respectively before CLSM observation.
For analysis of DR5::GUS activity, the 6-day-old seedlings were incubated with GUS staining solution at 37°C for 1 hr and observed under the microscope. Images were processed using Image J software and Adobe Photoshop CS3.
Supporting Information
Rhizoids and callus induced by cytokinin and auxin. Effect of various combinations of 2, 4-D and kinetin on the formation of rhizoids and callus from primary leaf explants. Scale bar, 1 cm.
(TIF)
Analysis of RcRR1 transcript in different tissues. The expression of RcRR1 in roots (R), shoots (S), leaves (L), and flowers (F) was analyzed by real-time PCR. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 cycles.
(TIF)
Analysis of RcRR1 transcript during rhizoid development. The expression of RcRR1 in leaflets (LE), callus-early (CE), callus-mid (CM), callus-late (CL), and rhizoids (RH) was anayzed by real-time PCR. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 cycles.
(TIF)
Expression of the RcRR1 gene under various stress conditions. Callus were subjected to the treatments of 20 µM gibberellin, 200 mM NaCl, 50 µM ABA, low temperature and 9 µM kinetin for 3 hr before collecting samples for RNA preparation. GA3, gibberellin; Na, NaCl; A, ABA; LT, low temperature; KT, kinetin; Un, untreated. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 and 30 cycles, respectively.
(TIF)
Cytokinin modulates the expression of a putative Rosa canina auxin efflux carrier gene. Expression of putative auxin efflux carrier gene (RcPIN) in explants incubated on medium containing 0, 0.5, 1.0, 1.5 and 2.0 µg/ml kinetin for 7 days. 18S rRNA was used as a control. Primers used for RT-PCR are listed in Table S1. 18S rRNA and putative RcPIN genes were amplified for 28 and 32 cycles, respectively.
(TIF)
Reduced sensitivity of RcRR1- OX Arabidopsis to cytokinin in primary root growth. Asterisks indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
(TIF)
Reduced cytokinin sensitivity of RcRR1- OX Arabidopsis in primary root elongation rate. Seedlings were grown for 3 days in 1/2 Murashige and Skoog (MS) medium and then transferred to the medium containing 0.2 µg/ml kinetin, and were grown for another 4 days before measurement of primary root length. Asterisks indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
(TIF)
List of primers used in this study.
(DOCX)
Acknowledgments
We thank Dr. P. Bruno Müller for providing the DR5::GFP plasmid.
Funding Statement
This research was supported by the Surface Project from National Natural Science Foundation of China (Grant No.31171993). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dubois A, Remay A, Raymond O, Balzergue S, Chauvet A, et al. (2011) Genomic approach to study floral development genes in Rosa spp. PLoS ONE 6: e28455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pati PK, Rath SP, Sharma M, Sood A, Ahuja PS (2006) In vitro propagation of rose–a review. Biotechnol Adv 24: 94–114. [DOI] [PubMed] [Google Scholar]
- 3. Pati PK, Kaur N, Sharma M, Ahuja PS (2010) In vitro propagation of rose. Methods Mol Biol 589: 163–176. [DOI] [PubMed] [Google Scholar]
- 4. Ibrahim R, Debergh PC (2001) Factors controlling high efficiency adventitious bud formation and plant regeneration from in vitro leaf explants of roses (Rosa hybrida L.). Scientia Horticulturae 88: 41–57. [Google Scholar]
- 5. Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, et al. (2011) Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev cell 21: 796–804. [DOI] [PubMed] [Google Scholar]
- 6. Tian C, Chen Y, Zhao X, Zhao L (2008) Plant regeneration through protocorm-like bodies induced from rhizoids using leaf explants of Rosa spp. Plant Cell Rep 27: 823–831. [DOI] [PubMed] [Google Scholar]
- 7. Miller CO, Skoog F, Okumura F, Von Saltza M, Strong F (1956) Isolation, Structure and Synthesis of Kinetin, a Substance Promoting Cell Division. J Am chem Soc 78: 1375–1380. [Google Scholar]
- 8. Zhao XY, Su YH, Cheng ZJ, Zhang XS (2008) Cell fate switch during in vitro plant organogenesis. J Integr Plant Biol 50: 816–824. [DOI] [PubMed] [Google Scholar]
- 9. El-Showk S, Ruonala R, Helariutta Y (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140: 1373–1383. [DOI] [PubMed] [Google Scholar]
- 10. Perilli S, Moubayidin L, Sabatini S (2010) The molecular basis of cytokinin function. Curr Opin Plant Biol 13: 21–26. [DOI] [PubMed] [Google Scholar]
- 11. Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana . Plant Cell 23: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, et al. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- 13. Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, et al. (2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis . Proc Natl Acad Sci USA 103: 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nieminen K, Immanen J, Laxell M, Kauppinen L, Tarkowski P, et al. (2008) Cytokinin signaling regulates cambial development in poplar. Proc Natl Acad Sci USA 105: 20032–20037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desikan R, Horák J, Chaban C, Mira-Rodado V, Witthöft J, et al. (2008) The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS ONE 3: e2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuppu S, Mishra N, Hu R, Sun L, Zhu X, et al. (2013) Water-deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS ONE 8: e64190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Werner T, Schmülling T (2009) Cytokinin action in plant development. Curr Opin Plant Biol 12: 527–538. [DOI] [PubMed] [Google Scholar]
- 18. Hwang I, Sheen J, Muller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380. [DOI] [PubMed] [Google Scholar]
- 19. Sakakibara H, Takei K (2002) Identification of cytokinin biosynthesis genes in Arabidopsis: a breakthrough for understanding the metabolic pathway and the regulation in higher plants. J Plant Growth Regul 21: 17–23. [DOI] [PubMed] [Google Scholar]
- 20. Sakakibara H (2005) Cytokinin biosynthesis and regulation. Vitam Horm 72: 271–287. [DOI] [PubMed] [Google Scholar]
- 21. Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, et al. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83. [DOI] [PubMed] [Google Scholar]
- 22. West AH, Stock AM (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26: 369–376. [DOI] [PubMed] [Google Scholar]
- 23. Muller B, Sheen J (2007) Advances in cytokinin signaling. Science 318: 68–69. [DOI] [PubMed] [Google Scholar]
- 24. Haberer G, Kieber JJ (2002) Cytokinins. New insights into a classic phytohormone. Plant Physiol 128: 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferreira FJ, Kieber JJ (2005) Cytokinin signaling. Curr Opin Plant Biol 8: 518–525. [DOI] [PubMed] [Google Scholar]
- 26. To JP, Kieber JJ (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92. [DOI] [PubMed] [Google Scholar]
- 27. Kiba T, Yamada H, Sato S, Kato T, Tabata S, et al. (2003) The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana . Plant Cell Physiol 44: 868–874. [DOI] [PubMed] [Google Scholar]
- 28. Mizuno T (2004) Plant response regulators implicated in signal transduction and circadian rhythm. Curr Opin Plant Biol 7: 499–505. [DOI] [PubMed] [Google Scholar]
- 29. Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, et al. (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis . Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiba T, Aoki K, Sakakibara H, Mizuno T (2004) Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol 45: 1063–1077. [DOI] [PubMed] [Google Scholar]
- 31. McClung CR (2006) Plant circadian rhythms. Plant Cell 18: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- 34. Barwale U, Kerns H, Widholm J (1986) Plant regeneration from callus cultures of several soybean genotypes via embryogenesis and organogenesis. Planta 167: 473–481. [DOI] [PubMed] [Google Scholar]
- 35. Ioio RD, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, et al. (2008) A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- 36. Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, et al. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682. [DOI] [PubMed] [Google Scholar]
- 37. Werner T, Motyka V, Strnad M, Schmülling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mira-Rodado V, Sweere U, Grefen C, Kunkel T, Fejes E, et al. (2007) Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis . J Exp Bot 58: 2595–2607. [DOI] [PubMed] [Google Scholar]
- 40. Choi J, Hwang I (2007) Cytokinin: perception, signal transduction, and role in plant growth and development. J Plant Biol 50: 98–108. [Google Scholar]
- 41. Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539. [DOI] [PubMed] [Google Scholar]
- 42. Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, et al. (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294: 1108–1111. [DOI] [PubMed] [Google Scholar]
- 43. To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, et al. (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis . Plant Cell 14: S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeon J, Kim NY, Kim S, Kang NY, Novák O, et al. (2010) A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis . J Biol Chem 285: 23371–23386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448. [DOI] [PubMed] [Google Scholar]
- 47. Camacho Y, Martínez-Castilla L, Fragoso S, Vazquez S, Martínez-Barajas E, et al. (2008) Characterization of a type A response regulator in the common bean (Phaseolus vulgaris) in response to phosphate starvation. Physiol Plant 132: 272–282. [DOI] [PubMed] [Google Scholar]
- 48. Zhang S, Lemaux PG (2004) Molecular analysis of in vitro shoot organogenesis. Crit Rev Plant Sci 23: 325–335. [Google Scholar]
- 49. Sugiyama M (1999) Organogenesis in vitro . Curr Opin Plant Biol 2: 61–64. [DOI] [PubMed] [Google Scholar]
- 50. Christianson M, Warnick D (1985) Temporal requirement for phytohormone balance in the control of organogenesis in vitro . Dev Biol 112: 494–497. [Google Scholar]
- 51. Pernisová M, Klíma P, Horák J, Válková M, Malbeck J, et al. (2009) Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc Natl Acad Sci USA 106: 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 53. Batoko H, Zheng H-Q, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rhizoids and callus induced by cytokinin and auxin. Effect of various combinations of 2, 4-D and kinetin on the formation of rhizoids and callus from primary leaf explants. Scale bar, 1 cm.
(TIF)
Analysis of RcRR1 transcript in different tissues. The expression of RcRR1 in roots (R), shoots (S), leaves (L), and flowers (F) was analyzed by real-time PCR. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 cycles.
(TIF)
Analysis of RcRR1 transcript during rhizoid development. The expression of RcRR1 in leaflets (LE), callus-early (CE), callus-mid (CM), callus-late (CL), and rhizoids (RH) was anayzed by real-time PCR. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 cycles.
(TIF)
Expression of the RcRR1 gene under various stress conditions. Callus were subjected to the treatments of 20 µM gibberellin, 200 mM NaCl, 50 µM ABA, low temperature and 9 µM kinetin for 3 hr before collecting samples for RNA preparation. GA3, gibberellin; Na, NaCl; A, ABA; LT, low temperature; KT, kinetin; Un, untreated. 18S rRNA was used as a control. 18S rRNA and RcRR1 genes were amplified for 28 and 30 cycles, respectively.
(TIF)
Cytokinin modulates the expression of a putative Rosa canina auxin efflux carrier gene. Expression of putative auxin efflux carrier gene (RcPIN) in explants incubated on medium containing 0, 0.5, 1.0, 1.5 and 2.0 µg/ml kinetin for 7 days. 18S rRNA was used as a control. Primers used for RT-PCR are listed in Table S1. 18S rRNA and putative RcPIN genes were amplified for 28 and 32 cycles, respectively.
(TIF)
Reduced sensitivity of RcRR1- OX Arabidopsis to cytokinin in primary root growth. Asterisks indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
(TIF)
Reduced cytokinin sensitivity of RcRR1- OX Arabidopsis in primary root elongation rate. Seedlings were grown for 3 days in 1/2 Murashige and Skoog (MS) medium and then transferred to the medium containing 0.2 µg/ml kinetin, and were grown for another 4 days before measurement of primary root length. Asterisks indicate statistically significant differences (Student’s t-test; P<0.01) between the control and transgenic plants; error bars show SDs.
(TIF)
List of primers used in this study.
(DOCX)