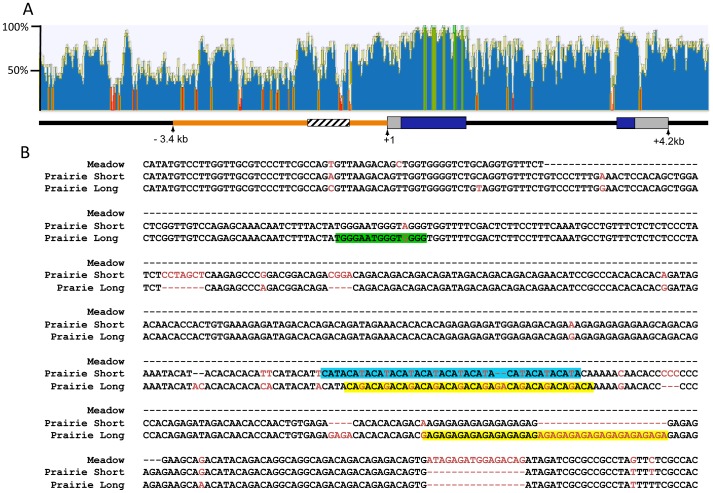
Figure 1. Comparison of the mouse and vole Avpr1a locus.
ClustalW was used to align 10.8 kb of mouse and prairie vole sequence containing the Avpr1a gene and sequence identity was calculated using a sliding 30 bp window using Geneious software (A). Green indicates areas of 100% identical sequence while red areas have <30% sequence identity between vole and mouse. The microsatellite region is shown in cross hatch and the 5′ flanking region targeted for replacement is shown in orange. The pairwise percent identity of the replaced 5′ flanking region between prairie vole and mouse is 53.6%. (B) Voles have a complex microsatellite element upstream of Avpr1a (crosshatched region; A) that exhibits both species and individual differences in sequence composition and length. The alignment of the meadow and prairie microsatellite alleles used in our targeting vectors is shown. Sequence differences are shown in red, and potential differential transcription factor bindings sites have been shaded. Green = Rreb1 binding site unique to the long allele; blue and yellow regions indicate differential binding opportunities for factors recognizing TATA-like and GAGA-like sequences, respectively. Although not shown, the montane vole has the same general structure as the meadow vole with regard to the microsatellite.