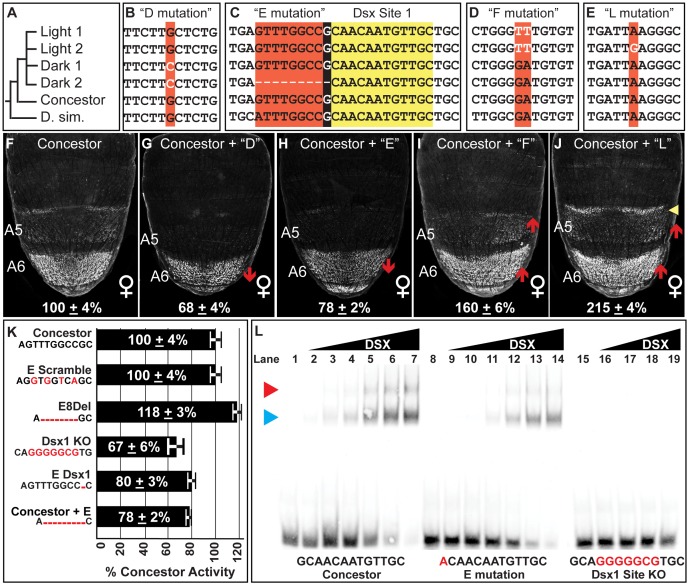
Figure 5. Functionally-relevant mutations in dimorphic element alleles.
(A) Dimorphic element allele phylogeny, including the outgroup species D. simulans (D. sim.). Alignment of sequences encompassing the (B) “D” mutation, (C) “E” mutation, (D) “F” mutation, (E) and the “L” mutation. Black background color for the E mutation indicates the 1 base pair overlap for the derived deletion and the adjacent DSX binding site. (F–J) Comparison of GFP-reporter activity in female transgenic pupae at 85 hAPF, represented as the % of the D. melanogaster Concestor element female A6 mean ± SEM. Red upward and downward arrow respectively indicate segments with increased and decreased regulatory activity. Yellow arrowhead indicates expanded regulatory activity. Regulatory activities differing from the Concestor element due to the following derived mutations: (G) D mutation; (H) E mutation; (I) F mutation; and (J) L mutation. (K) Summary for the female A6 regulatory activities for modifications to the E mutation region. The Concestor element sequence is provided and the introduced modifications indicated by red bases. (L) Gel shift assays for annealed oligonucleotide probes containing the wild type (Concestor element, lanes 1–7), E mutation (lanes 8–14), and mutant (Dsx1 KO, lanes 15–19) Dsx1 binding site. The binding site sequences are included with mutant bases in red. For the Concestor element and E mutation probes, binding reactions used increasing amounts of the DSX protein (from left to right: 0 ng, 8 ng, 16 ng, 31 ng, 63 ng, 125 ng, 250 ng, and 500 ng). For the Dsx1 KO probe, binding reactions used the following amounts of protein (from left to right: 0 ng, 8 ng, 31 ng, 125 ng, 500 ng). Blue and red arrowheads point to the respective locations of single or pair of DSX monomers bound to the probe.