Figure 2. Data flow.
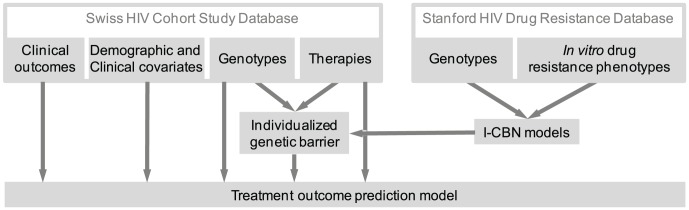
Matched pairs of viral genotype and drug resistance phenotype from the Stanford HIV Drug Resistance Database (top right) were used to learn I-CBN models for all drugs separately. The drug-specific individualized genetic barriers (IGBs) are derived from these models. The IGB to regimen is computed for each genotype-therapy pair in the Swiss HIV Cohort Database and its predictive power is assessed in prediction models that also account for classical demographic, clinical, and genetic covariates.
